A representation learning model based on variational inference and graph autoencoder for predicting lncRNA-disease associations
- PMID: 33745450
- PMCID: PMC7983260
- DOI: 10.1186/s12859-021-04073-z
A representation learning model based on variational inference and graph autoencoder for predicting lncRNA-disease associations
Abstract
Background: Numerous studies have demonstrated that long non-coding RNAs are related to plenty of human diseases. Therefore, it is crucial to predict potential lncRNA-disease associations for disease prognosis, diagnosis and therapy. Dozens of machine learning and deep learning algorithms have been adopted to this problem, yet it is still challenging to learn efficient low-dimensional representations from high-dimensional features of lncRNAs and diseases to predict unknown lncRNA-disease associations accurately.
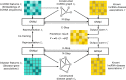
Results: We proposed an end-to-end model, VGAELDA, which integrates variational inference and graph autoencoders for lncRNA-disease associations prediction. VGAELDA contains two kinds of graph autoencoders. Variational graph autoencoders (VGAE) infer representations from features of lncRNAs and diseases respectively, while graph autoencoders propagate labels via known lncRNA-disease associations. These two kinds of autoencoders are trained alternately by adopting variational expectation maximization algorithm. The integration of both the VGAE for graph representation learning, and the alternate training via variational inference, strengthens the capability of VGAELDA to capture efficient low-dimensional representations from high-dimensional features, and hence promotes the robustness and preciseness for predicting unknown lncRNA-disease associations. Further analysis illuminates that the designed co-training framework of lncRNA and disease for VGAELDA solves a geometric matrix completion problem for capturing efficient low-dimensional representations via a deep learning approach.
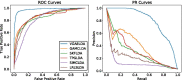
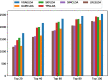
Conclusion: Cross validations and numerical experiments illustrate that VGAELDA outperforms the current state-of-the-art methods in lncRNA-disease association prediction. Case studies indicate that VGAELDA is capable of detecting potential lncRNA-disease associations. The source code and data are available at https://github.com/zhanglabNKU/VGAELDA .
Keywords: Graph autoencoder; Representation learning; Variational inference; lncRNA-disease association.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
Attentional multi-level representation encoding based on convolutional and variance autoencoders for lncRNA-disease association prediction.Brief Bioinform. 2021 May 20;22(3):bbaa067. doi: 10.1093/bib/bbaa067. Brief Bioinform. 2021. PMID: 32444875
-
GAE-LGA: integration of multi-omics data with graph autoencoders to identify lncRNA-PCG associations.Brief Bioinform. 2022 Nov 19;23(6):bbac452. doi: 10.1093/bib/bbac452. Brief Bioinform. 2022. PMID: 36305456
-
Multi-channel graph attention autoencoders for disease-related lncRNAs prediction.Brief Bioinform. 2022 Mar 10;23(2):bbab604. doi: 10.1093/bib/bbab604. Brief Bioinform. 2022. PMID: 35108355
-
DeepWalk based method to predict lncRNA-miRNA associations via lncRNA-miRNA-disease-protein-drug graph.BMC Bioinformatics. 2022 Feb 25;22(Suppl 12):621. doi: 10.1186/s12859-022-04579-0. BMC Bioinformatics. 2022. PMID: 35216549 Free PMC article. Review.
-
Data resources and computational methods for lncRNA-disease association prediction.Comput Biol Med. 2023 Feb;153:106527. doi: 10.1016/j.compbiomed.2022.106527. Epub 2023 Jan 2. Comput Biol Med. 2023. PMID: 36610216 Review.
Cited by
-
Predicting miRNA-Disease Association Based on Neural Inductive Matrix Completion with Graph Autoencoders and Self-Attention Mechanism.Biomolecules. 2022 Jan 2;12(1):64. doi: 10.3390/biom12010064. Biomolecules. 2022. PMID: 35053212 Free PMC article.
-
IGCNSDA: unraveling disease-associated snoRNAs with an interpretable graph convolutional network.Brief Bioinform. 2024 Mar 27;25(3):bbae179. doi: 10.1093/bib/bbae179. Brief Bioinform. 2024. PMID: 38647155 Free PMC article.
-
LDAGM: prediction lncRNA-disease asociations by graph convolutional auto-encoder and multilayer perceptron based on multi-view heterogeneous networks.BMC Bioinformatics. 2024 Oct 15;25(1):332. doi: 10.1186/s12859-024-05950-z. BMC Bioinformatics. 2024. PMID: 39407120 Free PMC article.
-
Predicting lncRNA-disease associations using multiple metapaths in hierarchical graph attention networks.BMC Bioinformatics. 2024 Jan 29;25(1):46. doi: 10.1186/s12859-024-05672-2. BMC Bioinformatics. 2024. PMID: 38287236 Free PMC article.
-
Predicting noncoding RNA and disease associations using multigraph contrastive learning.Sci Rep. 2025 Jan 2;15(1):230. doi: 10.1038/s41598-024-81862-5. Sci Rep. 2025. PMID: 39747154 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

