Molecular Docking Reveals Ivermectin and Remdesivir as Potential Repurposed Drugs Against SARS-CoV-2
- PMID: 33746908
- PMCID: PMC7976659
- DOI: 10.3389/fmicb.2020.592908
Molecular Docking Reveals Ivermectin and Remdesivir as Potential Repurposed Drugs Against SARS-CoV-2
Abstract
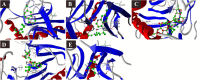
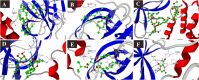
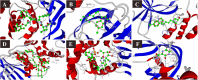
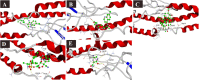
SARS-CoV-2 is a newly emerged coronavirus that causes a respiratory disease with variable severity and fatal consequences. It was first reported in Wuhan and subsequently caused a global pandemic. The viral spike protein binds with the ACE-2 cell surface receptor for entry, while TMPRSS2 triggers its membrane fusion. In addition, RNA dependent RNA polymerase (RdRp), 3'-5' exoribonuclease (nsp14), viral proteases, N, and M proteins are important in different stages of viral replication. Accordingly, they are attractive targets for different antiviral therapeutic agents. Although many antiviral agents have been used in different clinical trials and included in different treatment protocols, the mode of action against SARS-CoV-2 is still not fully understood. Different potential repurposed drugs, including, chloroquine, hydroxychloroquine, ivermectin, remdesivir, and favipiravir, were screened in the present study. Molecular docking of these drugs with different SARS-CoV-2 target proteins, including spike and membrane proteins, RdRp, nucleoproteins, viral proteases, and nsp14, was performed. Moreover, the binding affinities of the human ACE-2 receptor and TMPRSS2 to the different drugs were evaluated. Molecular dynamics simulation and MM-PBSA calculation were also conducted. Ivermectin and remdesivir were found to be the most promising drugs. Our results suggest that both these drugs utilize different mechanisms at the entry and post-entry stages and could be considered potential inhibitors of SARS-CoV-2 replication.
Keywords: COVID-19; SARS-CoV-2; antiviral; chloroquine/hydroxychloroquine; coronavirus disease; favipiravir; ivermectin; remdesivir (GS-5734).
Copyright © 2021 Eweas, Alhossary and Abdel-Moneim.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Abraham M. J., Murtola T., Schulz R., Páll S., Smith J. C., Hess B., et al. (2015). GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1 19–25. 10.1016/j.softx.2015.06.001 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

