The Surge of Hypervirulent ST398 MRSA Lineage With Higher Biofilm-Forming Ability Is a Critical Threat to Clinics
- PMID: 33746929
- PMCID: PMC7969815
- DOI: 10.3389/fmicb.2021.636788
The Surge of Hypervirulent ST398 MRSA Lineage With Higher Biofilm-Forming Ability Is a Critical Threat to Clinics
Abstract
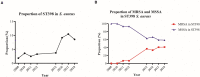
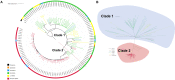
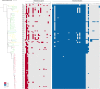
The global increase of community-associated (CA) infections with methicillin-resistant Staphylococcus aureus (MRSA) is a major healthcare problem. Although sequence type (ST) 398 MRSA was first described as a livestock-associated (LA) lineage, human-adapted MRSA (HO-MRSA) ST398 without livestock contact has subsequently been reported from China in our previous study and other later research. The proportion of ST398 HO-MRSA has also remarkably increased in recent years in China. Based on 3878 S. aureus isolates that were collected in a general hospital between 2008 and 2018, we identified 56 ST398 HO-MRSA isolates. The four early appearing isolates of them have been sequenced by whole-genome sequencing (WGS) in our previous study. Here, by usage of WGS on the later-appearing 52 isolates and analyzing the phylogenetic dynamics of the linage, we found that 50 isolates clustered together with the former 4 isolates, making it a main clade out of MSSA clones and other MRSA clones, although ST398 HO-MRSA evolved with multiple origins. Drug resistance and virulence gene analysis based on the WGS data demonstrated that ST398 HO-MRSA main clade exhibited a similar pattern in both parts. Furthermore, they all carried a conserved variant of prophage 3 to guarantee virulence and a short SCCmec type V element of class D to maintain considerable lower methicillin resistance. Further phenotypical research verified that the epidemic HO-MRSA ST398 displayed enhanced biofilm formation ability when keeping high virulence. The dual advantages of virulence and biofilm formation in the HO-MRSA ST398 subtype promote their fitness in the community and even in the healthcare environment, which poses a serious threat in clinical S. aureus infections. Therefore, further surveillance is required to prevent and control the problematic public health impact of HO-MRSA ST398 in the future.
Keywords: methicillin-resistant staphylococcus aureus; phylogenetic analysis; sequence type 398; virulence; whole-genome sequencing.
Copyright © 2021 Lu, Zhao, Si, Jian, Wang, Li, Dai, Huang, Ma, He and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Dai Y., Liu J., Guo W., Meng H., Huang Q., He L., et al. (2019). Decreasing methicillin-resistant Staphylococcus aureus (MRSA) infections is attributable to the disappearance of predominant MRSA ST239 clones, Shanghai, 2008-2017. Emerg. Microbes. Infect. 8 471–478. 10.1080/22221751.2019.1595161 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

