Proteomic Analysis of Fusarium oxysporum-Induced Mechanism in Grafted Watermelon Seedlings
- PMID: 33747013
- PMCID: PMC7969889
- DOI: 10.3389/fpls.2021.632758
Proteomic Analysis of Fusarium oxysporum-Induced Mechanism in Grafted Watermelon Seedlings
Abstract
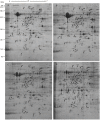
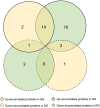
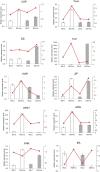
Grafting can improve the resistance of watermelon to soil-borne diseases. However, the molecular mechanism of defense response is not completely understood. Herein, we used a proteomic approach to investigate the molecular basis involved in grafted watermelon leaf defense against Fusarium oxysporum f.sp. niveum (FON) infection. The bottle gourd rootstock-grafted (RG) watermelon seedlings were highly resistant to FON compared with self-grafted (SG) watermelon plants, with a disease incidence of 3.4 and 89%, respectively. Meanwhile, grafting significantly induced the activity of pathogenesis-related proteases under FON challenge. Proteins extracted from leaves of RG and SG under FON inoculation were analyzed using two-dimensional gel electrophoresis. Thirty-nine differentially accumulated proteins (DAPs) were identified and classified into 10 functional groups. Accordingly, protein biosynthetic and stress- and defense-related proteins play crucial roles in the enhancement of disease resistance of RG watermelon seedlings, compared with that of SG watermelon seedlings. Proteins involved in signal transduction positively regulated the defense process. Carbohydrate and energy metabolism and photosystem contributed to energy production in RG watermelon seedlings under FON infection. The disease resistance of RG watermelon seedlings may also be related to the improved scavenging capacity of reactive oxygen species (ROS). The expression profile of 10 randomly selected proteins was measured using quantitative real-time PCR, among which, 7 was consistent with the results of the proteomic analysis. The functional implications of these proteins in regulating grafted watermelon response against F. oxysporum are discussed.
Keywords: Citrullus lanatus; Fusarium oxysporum f.sp. niveum; bottle gourd; proteomics; rootstock grafting.
Copyright © 2021 Zhang, Xu, Ren, Liu, Yao, Lou, Xu and Yang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
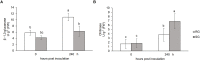


References
-
- Beracochea V. C., Almasia N. I., Peluffo L., Nahirnak V., Hopp E. H., Paniego N., et al. (2015). Sunflower germin-like protein HaGLP1 promotes ROS accumulation and enhances protection against fungal pathogens in transgenic Arabidopsis thaliana. Plant Cell. Rep. 34 1717–1733. 10.1007/s00299-015-1819-4 - DOI - PubMed
-
- Berger S., Papadopoulos M., Schreiber U., Kaiser W., Roitsch T. (2004). Complex regulation of gene expression, photosynthesis and sugar levels by pathogen infection in tomato. Physiol. Plantarum. 122, 419–428. 10.1111/j.1399-3054.2004.00433.x - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

