Prognostic Risk Model of Immune-Related Genes in Colorectal Cancer
- PMID: 33747044
- PMCID: PMC7970128
- DOI: 10.3389/fgene.2021.619611
Prognostic Risk Model of Immune-Related Genes in Colorectal Cancer
Abstract
Purpose: We focused on immune-related genes (IRGs) derived from transcriptomic studies, which had the potential to stratify patients' prognosis and to establish a risk assessment model in colorectal cancer.
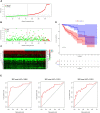
Summary: This article examined our understanding of the molecular pathways associated with intratumoral immune response, which represented a critical step for the implementation of stratification strategies toward the development of personalized immunotherapy of colorectal cancer. More and more evidence shows that IRGs play an important role in tumors. We have used data analysis to screen and identify immune-related molecular biomarkers of colon cancer. We selected 18 immune-related prognostic genes and established models to assess prognostic risks of patients, which can provide recommendations for clinical treatment and follow-up. Colorectal cancer (CRC) is a leading cause of cancer-related death in human. Several studies have investigated whether IRGs and tumor immune microenvironment (TIME) could be indicators of CRC prognoses. This study aimed to develop an improved prognostic signature for CRC based on IRGs to predict overall survival (OS) and provide new therapeutic targets for CRC treatment. Based on the screened IRGs, the Cox regression model was used to build a prediction model based on 18-IRG signature. Cox regression analysis revealed that the 18-IRG signature was an independent prognostic factor for OS in CRC patients. Then, we used the TIMER online database to explore the relationship between the risk scoring model and the infiltration of immune cells, and the results showed that the risk model can reflect the state of TIME to a certain extent. In short, an 18-IRG prognostic signature for predicting CRC patients' survival was firmly established.
Keywords: TCGA; colorectal cancer; immune prognostic signature; immune-related gene; tumor immune microenvironment.
Copyright © 2021 Qian, Wei, Lu, Sun, Hwang, Jiang, Fu, Zhou, Kong, Zhu, Xiao, Hu and Ding.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures







References
LinkOut - more resources
Full Text Sources
Other Literature Sources

