Variation, Variegation and Heritable Gene Repression in S. cerevisiae
- PMID: 33747046
- PMCID: PMC7970126
- DOI: 10.3389/fgene.2021.630506
Variation, Variegation and Heritable Gene Repression in S. cerevisiae
Abstract
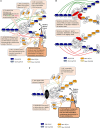
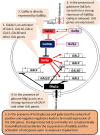
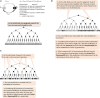
Phenotypic heterogeneity provides growth advantages for a population upon changes of the environment. In S. cerevisiae, such heterogeneity has been observed as "on/off" states in the expression of individual genes in individual cells. These variations can persist for a limited or extended number of mitotic divisions. Such traits are known to be mediated by heritable chromatin structures, by the mitotic transmission of transcription factors involved in gene regulatory circuits or by the cytoplasmic partition of prions or other unstructured proteins. The significance of such epigenetic diversity is obvious, however, we have limited insight into the mechanisms that generate it. In this review, we summarize the current knowledge of epigenetically maintained heterogeneity of gene expression and point out similarities and converging points between different mechanisms. We discuss how the sharing of limiting repression or activation factors can contribute to cell-to-cell variations in gene expression and to the coordination between short- and long- term epigenetic strategies. Finally, we discuss the implications of such variations and strategies in adaptation and aging.
Keywords: chromatin; diversity; gene regulatory circuits; gene repression; gene silencing; long non-coding RNA; phenotypic heterogeneity.
Copyright © 2021 Shaban, Sauty and Yankulov.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

