The Roles of CircRNAs in Regulating Muscle Development of Livestock Animals
- PMID: 33748107
- PMCID: PMC7973088
- DOI: 10.3389/fcell.2021.619329
The Roles of CircRNAs in Regulating Muscle Development of Livestock Animals
Abstract
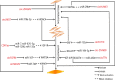
The muscle growth and development of livestock animals is a complex, multistage process, which is regulated by many factors, especially the genes related to muscle development. In recent years, it has been reported frequently that circular RNAs (circRNAs) are involved widely in cell proliferation, cell differentiation, and body development (including muscle development). However, the research on circRNAs in muscle growth and development of livestock animals is still in its infancy. In this paper, we briefly introduce the discovery, classification, biogenesis, biological function, and degradation of circRNAs and focus on the molecular mechanism and mode of action of circRNAs as competitive endogenous RNAs in the muscle development of livestock and poultry. In addition, we also discuss the regulatory mechanism of circRNAs on muscle development in livestock in terms of transcription, translation, and mRNAs. The purpose of this article is to discuss the multiple regulatory roles of circRNAs in the process of muscle development in livestock, to provide new ideas for the development of a new co-expression regulation network, and to lay a foundation for enriching livestock breeding and improving livestock economic traits.
Keywords: circRNAs; co-expression regulatory network; livestock animals; muscle development; transcription and translation.
Copyright © 2021 Yang, He and Chen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Biological functions of circRNAs and their progress in livestock and poultry.Reprod Domest Anim. 2020 Dec;55(12):1667-1677. doi: 10.1111/rda.13816. Epub 2020 Oct 14. Reprod Domest Anim. 2020. PMID: 32916770 Review.
-
Functional Role of circRNAs in the Regulation of Fetal Development, Muscle Development, and Lactation in Livestock.Biomed Res Int. 2021 Feb 19;2021:5383210. doi: 10.1155/2021/5383210. eCollection 2021. Biomed Res Int. 2021. PMID: 33688493 Free PMC article. Review.
-
The emerging roles of circRNAs in traits associated with livestock breeding.Wiley Interdiscip Rev RNA. 2023 Jul-Aug;14(4):e1775. doi: 10.1002/wrna.1775. Epub 2023 Jan 11. Wiley Interdiscip Rev RNA. 2023. PMID: 36631071 Review.
-
Expression profiles of circular RNAs in sheep skeletal muscle.Asian-Australas J Anim Sci. 2018 Oct;31(10):1550-1557. doi: 10.5713/ajas.17.0563. Epub 2018 Apr 11. Asian-Australas J Anim Sci. 2018. PMID: 29642686 Free PMC article.
-
The emerging role of circRNAs on skeletal muscle development in economical animals.Anim Biotechnol. 2023 Dec;34(7):2778-2792. doi: 10.1080/10495398.2022.2118130. Epub 2022 Sep 2. Anim Biotechnol. 2023. PMID: 36052979 Review.
Cited by
-
Biological functions of circRNAs and their advance on skeletal muscle development in bovine.3 Biotech. 2023 May;13(5):133. doi: 10.1007/s13205-023-03558-3. Epub 2023 Apr 21. 3 Biotech. 2023. PMID: 37096117 Free PMC article. Review.
-
The circular RNA circNFIX regulates MEF2C expression in muscle satellite cells in spastic cerebral palsy.J Biol Chem. 2024 Dec;300(12):107987. doi: 10.1016/j.jbc.2024.107987. Epub 2024 Nov 13. J Biol Chem. 2024. PMID: 39542245 Free PMC article.
-
Transcriptomic Analysis Reveals Differentially Expressed Circular RNAs Associated with Fecundity in the Sheep Hypothalamus with Different FecB Genotypes.Animals (Basel). 2024 Jan 7;14(2):198. doi: 10.3390/ani14020198. Animals (Basel). 2024. PMID: 38254366 Free PMC article.
-
Editorial: Role of Non-Coding RNAs in Animals.Animals (Basel). 2023 Feb 23;13(5):805. doi: 10.3390/ani13050805. Animals (Basel). 2023. PMID: 36899662 Free PMC article.
-
Combined analysis of lncRNA and mRNA emphasizes the potential role of tryptophan-mediated regulation of muscle development in weaned piglets by lncRNA.J Anim Sci. 2024 Jan 3;102:skae264. doi: 10.1093/jas/skae264. J Anim Sci. 2024. PMID: 39276131 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

