A Novel Nine Apoptosis-Related Genes Signature Predicting Overall Survival for Kidney Renal Clear Cell Carcinoma and its Associations with Immune Infiltration
- PMID: 33748185
- PMCID: PMC7969794
- DOI: 10.3389/fmolb.2021.567730
A Novel Nine Apoptosis-Related Genes Signature Predicting Overall Survival for Kidney Renal Clear Cell Carcinoma and its Associations with Immune Infiltration
Abstract
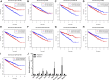
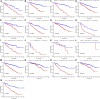
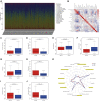
Background: This study was designed to establish a sensitive prognostic model based on apoptosis-related genes to predict overall survival (OS) in patients with clear cell renal cell carcinoma (ccRCC). Methods: Obtaining the expression of apoptosis-related genes and associated clinical parameters from online datasets (The Cancer Genome Atlas, TCGA), their biological function analyses were performed through differently expressed genes. By means of LASSO, unadjusted and adjusted Cox regression analyses, this predictive signature was constructed and validated by internal and external databases (both TCGA and ArrayExpress). Results: A total of nine apoptosis-related genes (SLC27A2, TNFAIP2, IFI44, CSF2, IL4, MDK, DOCK8, WNT5A, APP) were ultimately screened as associated hub genes and utilized to construct a prognosis model. Then our constructed riskScore model significantly passed the validation in both the internal and external datasets of OS (all p < 0.05) and verified their expressions by qRT-PCR. Moreover, we conducted the Receiver Operating Characteristic (ROC), finding the area under the ROC curves (AUCs) were all above 0.70 which indicated that riskScore was a stable independent prognostic factor (p < 0.05). Furthermore, prognostic nomograms were established to figure out the relationship between 1-, 3- and 5-year OS and individual parameters for ccRCC patients. Additionally, survival analyses indicated that our riskScore worked well in predicting OS in subgroups of age, gender, grade, stage, T, M, N0, White (all p < 0.05), except for African, Asian and N1 (p > 0.05). We also explored its association with immune infiltration and applied cMap database to seek out highly correlated small molecule drugs. Conclusion: Our study successfully constructed a prognostic model containing nine hub apoptosis-related genes for ccRCC, helping clinicians predict patients' OS and making the prognostic assessment more standardized. Future prospective studies are required to validate our findings.
Keywords: apoptosis-related genes; clear cell renal cell carcinoma; overall survival; prognosis; signature.
Copyright © 2021 Wang, Chen, Zhu, Ma and Xing.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures







References
LinkOut - more resources
Full Text Sources
Other Literature Sources

