Rad52 SUMOylation functions as a molecular switch that determines a balance between the Rad51- and Rad59-dependent survivors
- PMID: 33748714
- PMCID: PMC7966982
- DOI: 10.1016/j.isci.2021.102231
Rad52 SUMOylation functions as a molecular switch that determines a balance between the Rad51- and Rad59-dependent survivors
Abstract
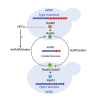
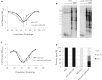
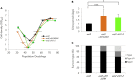
Functional telomeres in yeast lacking telomerase can be restored by rare Rad51- or Rad59-dependent recombination events that lead to type I and type II survivors, respectively. We previously proposed that polySUMOylation of proteins and the SUMO-targeted ubiquitin ligase Slx5-Slx8 are key factors in type II recombination. Here, we show that SUMOylation of Rad52 favors the formation of type I survivors. Conversely, preventing Rad52 SUMOylation partially bypasses the requirement of Slx5-Slx8 for type II recombination. We further report that SUMO-dependent proteasomal degradation favors type II recombination. Finally, inactivation of Rad59, but not Rad51, impairs the relocation of eroded telomeres to the Nuclear Pore complexes (NPCs). We propose that Rad59 cooperates with non-SUMOylated Rad52 to promote type II recombination at NPCs, resulting in the emergence of more robust survivors akin to ALT cancer cells. Finally, neither Rad59 nor Rad51 is required by itself for the survival of established type II survivors.
Keywords: Biological Sciences; Cell Biology; Molecular Biology.
© 2021 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Bergink S., Ammon T., Kern M., Schermelleh L., Leonhardt H., Jentsch S. Role of Cdc48/p97 as a SUMO-targeted segregase curbing Rad51-Rad52 interaction. Nat. Cell Biol. 2013;15:526–532. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

