Identifying CNS-colonizing T cells as potential therapeutic targets to prevent progression of multiple sclerosis
- PMID: 33748804
- PMCID: PMC7966680
- DOI: 10.1016/j.medj.2021.01.006
Identifying CNS-colonizing T cells as potential therapeutic targets to prevent progression of multiple sclerosis
Abstract
Background: Multiple sclerosis (MS), an autoimmune disease of the central nervous system (CNS), can be suppressed in its early stages but eventually becomes clinically progressive and unresponsive to therapy. Here, we investigate whether the therapeutic resistance of progressive MS can be attributed to chronic immune cell accumulation behind the blood-brain barrier (BBB).
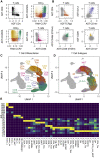
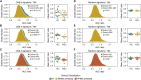
Methods: We systematically track CNS-homing immune cells in the peripheral blood of 31 MS patients and 31 matched healthy individuals in an integrated analysis of 497,705 single-cell transcriptomes and 355,433 surface protein profiles from 71 samples. Through spatial RNA sequencing, we localize these cells in post mortem brain tissue of 6 progressive MS patients contrasted against 4 control brains (20 samples, 85,000 spot transcriptomes).
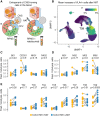
Findings: We identify a specific pathogenic CD161+/lymphotoxin beta (LTB)+ T cell population that resides in brains of progressive MS patients. Intriguingly, our data suggest that the colonization of the CNS by these T cells may begin earlier in the disease course, as they can be mobilized to the blood by usage of the integrin-blocking antibody natalizumab in relapsing-remitting MS patients.
Conclusions: As a consequence, we lay the groundwork for a therapeutic strategy to deplete CNS-homing T cells before they can fuel treatment-resistant progression.
Funding: This study was supported by funding from the University Medical Center Hamburg-Eppendorf, the Stifterverband für die Deutsche Wissenschaft, the OAK Foundation, Medical Research Council UK, and Wellcome.
Keywords: CNS-homing; T cells; Tfh; Th17; multiple sclerosis; natalizumab; scRNA-seq; spatial transcriptomics; therapeutic resistance.
© 2021 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





Comment in
-
Capturing pathogenic immune cells before they home to brain.Med. 2021 Mar 12;2(3):214-216. doi: 10.1016/j.medj.2021.02.005. Med. 2021. PMID: 33796875 Free PMC article.
References
-
- Dendrou C.A., Fugger L., Friese M.A. Immunopathology of multiple sclerosis. Nat. Rev. Immunol. 2015;15:545–558. - PubMed
-
- Polman C.H., O’Connor P.W., Havrdova E., Hutchinson M., Kappos L., Miller D.H., Phillips J.T., Lublin F.D., Giovannoni G., Wajgt A., AFFIRM Investigators A randomized, placebo-controlled trial of natalizumab for relapsing multiple sclerosis. N. Engl. J. Med. 2006;354:899–910. - PubMed
-
- Kapoor R., Ho P.-R., Campbell N., Chang I., Deykin A., Forrestal F., Lucas N., Yu B., Arnold D.L., Freedman M.S., ASCEND investigators Effect of natalizumab on disease progression in secondary progressive multiple sclerosis (ASCEND): a phase 3, randomised, double-blind, placebo-controlled trial with an open-label extension. Lancet Neurol. 2018;17:405–415. - PubMed
-
- Scalfari A., Romualdi C., Nicholas R.S., Mattoscio M., Magliozzi R., Morra A., Monaco S., Muraro P.A., Calabrese M. The cortical damage, early relapses, and onset of the progressive phase in multiple sclerosis. Neurology. 2018;90:e2107–e2118. - PubMed
-
- Magliozzi R., Howell O.W., Reeves C., Roncaroli F., Nicholas R., Serafini B., Aloisi F., Reynolds R. A Gradient of neuronal loss and meningeal inflammation in multiple sclerosis. Ann. Neurol. 2010;68:477–493. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

