Bioinformatics-driven discovery of novel Clostridioides difficile lysins and experimental comparison with highly active benchmarks
- PMID: 33748952
- PMCID: PMC10049856
- DOI: 10.1002/bit.27759
Bioinformatics-driven discovery of novel Clostridioides difficile lysins and experimental comparison with highly active benchmarks
Abstract
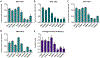
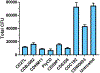
Clostridioides difficile is the single most deadly bacterial pathogen in the United States, and its global prevalence and outsized health impacts underscore the need for more effective therapeutic options. Towards this goal, a novel group of modified peptidoglycan hydrolases with significant in vitro bactericidal activity have emerged as potential candidates for treating C. difficile infections (CDI). To date, discovery and development efforts directed at these CDI-specific lysins have been limited, and in particular there has been no systematic comparison of known or newly discovered lysin candidates. Here, we detail bioinformatics-driven discovery of six new anti-C. difficile lysins belonging to the amidase-3 family of enzymes, and we describe experimental comparison of their respective catalytic domains (CATs) with highly active CATs from the literature. Our quantitative analyses include metrics for expression level, inherent antibacterial activity, breadth of strain selectivity, killing of germinating spores, and structural and functional measures of thermal stability. Importantly, prior studies have not examined stability as a performance metric, and our results show that the panel of eight enzymes possess widely variable thermal denaturation temperatures and resistance to heat inactivation, including some enzymes that exhibit marginal stability at body temperature. Ultimately, no single enzyme dominated with respect to all performance measures, suggesting the need for a balanced assessment of lysin properties during efforts to find, engineer, and develop candidates with true clinical potential.
Keywords: C. difficile; antibiotic; autolysin; peptidoglycan hydrolase; thermostability.
© 2021 Wiley Periodicals LLC.
Conflict of interest statement
Competing Interests
KEG and CB-K are co-founders of Lyticon LLC. No other authors have a conflict of interest. Potential conflicts of interest for KEG and CB-K are under management at Dartmouth. The authors declare that the work presented here is free of any bias.
Figures



Similar articles
-
Dimer-monomer transition defines a hyper-thermostable peptidoglycan hydrolase mined from bacterial proteome by lysin-derived antimicrobial peptide-primed screening.Elife. 2024 Nov 26;13:RP98266. doi: 10.7554/eLife.98266. Elife. 2024. PMID: 39589395 Free PMC article.
-
Structural and biochemical characterizations of the novel autolysin Acd24020 from Clostridioides difficile and its full-function catalytic domain as a lytic enzyme.Mol Microbiol. 2021 Apr;115(4):684-698. doi: 10.1111/mmi.14636. Epub 2020 Nov 22. Mol Microbiol. 2021. PMID: 33140473
-
Ser/Thr Kinase-Dependent Phosphorylation of the Peptidoglycan Hydrolase CwlA Controls Its Export and Modulates Cell Division in Clostridioides difficile.mBio. 2021 May 18;12(3):e00519-21. doi: 10.1128/mBio.00519-21. mBio. 2021. PMID: 34006648 Free PMC article.
-
Clostridioides difficile peptidoglycan modifications.Curr Opin Microbiol. 2022 Feb;65:156-161. doi: 10.1016/j.mib.2021.11.010. Epub 2021 Dec 6. Curr Opin Microbiol. 2022. PMID: 34883390 Review.
-
Bacteriophage endolysins as a potential weapon to combat Clostridioides difficile infection.Gut Microbes. 2020 Nov 9;12(1):1813533. doi: 10.1080/19490976.2020.1813533. Gut Microbes. 2020. PMID: 32985336 Free PMC article. Review.
Cited by
-
Assessing the Feasibility of Employing a Combination of a Bacteriophage-Derived Endolysin and Spore Germinants to Treat Relapsing Clostridioides difficile Infection.Microorganisms. 2023 Jun 24;11(7):1651. doi: 10.3390/microorganisms11071651. Microorganisms. 2023. PMID: 37512824 Free PMC article.
References
-
- CDC. (2019). Antibiotic Resistance Threats in the United States, 2019. Centers for Disease Control; Retrieved from http://www.cdc.gov/drugresistance/Biggest-Threats.html
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

