Limited Predictability of Amino Acid Substitutions in Seasonal Influenza Viruses
- PMID: 33749787
- PMCID: PMC8233509
- DOI: 10.1093/molbev/msab065
Limited Predictability of Amino Acid Substitutions in Seasonal Influenza Viruses
Abstract
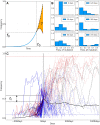
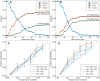
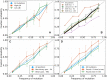
Seasonal influenza viruses repeatedly infect humans in part because they rapidly change their antigenic properties and evade host immune responses, necessitating frequent updates of the vaccine composition. Accurate predictions of strains circulating in the future could therefore improve the vaccine match. Here, we studied the predictability of frequency dynamics and fixation of amino acid substitutions. Current frequency was the strongest predictor of eventual fixation, as expected in neutral evolution. Other properties, such as occurrence in previously characterized epitopes or high Local Branching Index (LBI) had little predictive power. Parallel evolution was found to be moderately predictive of fixation. Although the LBI had little power to predict frequency dynamics, it was still successful at picking strains representative of future populations. The latter is due to a tendency of the LBI to be high for consensus-like sequences that are closer to the future than the average sequence. Simulations of models of adapting populations, in contrast, show clear signals of predictability. This indicates that the evolution of influenza HA and NA, while driven by strong selection pressure to change, is poorly described by common models of directional selection such as traveling fitness waves.
Keywords: evolution; influenza; population genetics.
© The Author(s) 2021. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures

References
-
- Bogner P, Capua I, Lipman DJ, Cox NJ, et al. 2006. A global initiative on sharing avian flu data. Nature 442(7106):981–981.
-
- Bush RM, Bender CA, Subbarao K, Cox NJ, Fitch WM.. 1999. Predicting the evolution of human influenza A. Science 286(5446):1921–1925. - PubMed
-
- Dunning I, Huchette J, Lubin M.. 2017. Jump: a modeling language for mathematical optimization. SIAM Rev. 59(2):295–320.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

