A small molecule compound berberine as an orally active therapeutic candidate against COVID-19 and SARS: A computational and mechanistic study
- PMID: 33749932
- PMCID: PMC8250068
- DOI: 10.1096/fj.202001792R
A small molecule compound berberine as an orally active therapeutic candidate against COVID-19 and SARS: A computational and mechanistic study
Abstract
The novel coronavirus disease, COVID-19, has grown into a global pandemic and a major public health threat since its breakout in December 2019. To date, no specific therapeutic drug or vaccine for treating COVID-19 and SARS has been FDA approved. Previous studies suggest that berberine, an isoquinoline alkaloid, has shown various biological activities that may help against COVID-19 and SARS, including antiviral, anti-allergy and inflammation, hepatoprotection against drug- and infection-induced liver injury, as well as reducing oxidative stress. In particular, berberine has a wide range of antiviral activities such as anti-influenza, anti-hepatitis C, anti-cytomegalovirus, and anti-alphavirus. As an ingredient recommended in guidelines issued by the China National Health Commission for COVID-19 to be combined with other therapy, berberine is a promising orally administered therapeutic candidate against SARS-CoV and SARS-CoV-2. The current study comprehensively evaluates the potential therapeutic mechanisms of berberine in preventing and treating COVID-19 and SARS using computational modeling, including target mining, gene ontology enrichment, pathway analyses, protein-protein interaction analysis, and in silico molecular docking. An orally available immunotherapeutic-berberine nanomedicine, named NIT-X, has been developed by our group and has shown significantly increased oral bioavailability of berberine, increased IFN-γ production by CD8+ T cells, and inhibition of mast cell histamine release in vivo, suggesting a protective immune response. We further validated the inhibition of replication of SARS-CoV-2 in lung epithelial cells line in vitro (Calu3 cells) by berberine. Moreover, the expression of targets including ACE2, TMPRSS2, IL-1α, IL-8, IL-6, and CCL-2 in SARS-CoV-2 infected Calu3 cells were significantly suppressed by NIT-X. By supporting protective immunity while inhibiting pro-inflammatory cytokines; inhibiting viral infection and replication; inducing apoptosis; and protecting against tissue damage, berberine is a promising candidate in preventing and treating COVID-19 and SARS. Given the high oral bioavailability and safety of berberine nanomedicine, the current study may lead to the development of berberine as an orally, active therapeutic against COVID-19 and SARS.
Keywords: COVID-19 and SARS; anti-viral; apoptosis; berberine; computational modeling.
© 2021 Federation of American Societies for Experimental Biology.
Conflict of interest statement
ZZW, YL, WH, RT, ANW, JG, and MM have no financial conflict of interests to disclose. XML received research support from the National Institutes of Health (NIH)/National Center for Complementary and Alternative Medicine (NCCAM); Food Allergy Research and Education (FARE) and Winston Wolkoff Integrative Medicine Fund for Allergies and Wellness; received consulting fees from Food Allergy Research and Education (FARE), Johnson & Joh nson Pharmaceutical Research & Development, LLC, Bayer Global Health LLC; Received grant from Henan University of Chinese Medicine & New York Medica College for TCM Immunopharmacology and Integrative Medicine; received royalties from UpToDate; received travel expenses from the National Center for Complementary and Alternative Medicine (NCCAM) and FARE; received practice compensation from the Integrative Health and Acupuncture PC; US Times Technology Inc is managed by the related party; Share patents (PCT/US14/857772, PCT/US14/68396, PCT/US2014/012306, PCT/US2005/008417, and (PCT/US2017/056822 (Pending), is a member of Herbs Springs, LLC, General Nutraceutical Technology LLC, and Health Freedom LLC. NY shares US patent PCT/US14/68396 and is a member of Health Freedom LLC. KS share patent (PCT/US2017/056822 (Pending), Salary support by General Nutraceutical Technology LLC.
Figures

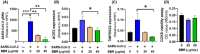
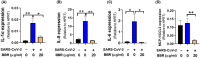
Similar articles
-
Computational exploration of the dual role of the phytochemical fortunellin: Antiviral activities against SARS-CoV-2 and immunomodulatory abilities against the host.Comput Biol Med. 2022 Oct;149:106049. doi: 10.1016/j.compbiomed.2022.106049. Epub 2022 Sep 8. Comput Biol Med. 2022. PMID: 36103744 Free PMC article.
-
Antiviral Activity of Type I, II, and III Interferons Counterbalances ACE2 Inducibility and Restricts SARS-CoV-2.mBio. 2020 Sep 10;11(5):e01928-20. doi: 10.1128/mBio.01928-20. mBio. 2020. PMID: 32913009 Free PMC article.
-
Comparative transcriptome analysis of SARS-CoV, MERS-CoV, and SARS-CoV-2 to identify potential pathways for drug repurposing.Comput Biol Med. 2021 Jan;128:104123. doi: 10.1016/j.compbiomed.2020.104123. Epub 2020 Nov 24. Comput Biol Med. 2021. PMID: 33260034 Free PMC article.
-
Nucleoside Analogs and Nucleoside Precursors as Drugs in the Fight against SARS-CoV-2 and Other Coronaviruses.Molecules. 2021 Feb 13;26(4):986. doi: 10.3390/molecules26040986. Molecules. 2021. PMID: 33668428 Free PMC article. Review.
-
The role of berberine in Covid-19: potential adjunct therapy.Inflammopharmacology. 2022 Dec;30(6):2003-2016. doi: 10.1007/s10787-022-01080-1. Epub 2022 Oct 2. Inflammopharmacology. 2022. PMID: 36183284 Free PMC article. Review.
Cited by
-
Research and development of Chinese anti-COVID-19 drugs.Acta Pharm Sin B. 2022 Dec;12(12):4271-4286. doi: 10.1016/j.apsb.2022.09.002. Epub 2022 Sep 13. Acta Pharm Sin B. 2022. PMID: 36119967 Free PMC article. Review.
-
Neuroprotective Agents with Therapeutic Potential for COVID-19.Biomolecules. 2023 Oct 27;13(11):1585. doi: 10.3390/biom13111585. Biomolecules. 2023. PMID: 38002267 Free PMC article. Review.
-
Assessing the Potential Contribution of In Silico Studies in Discovering Drug Candidates That Interact with Various SARS-CoV-2 Receptors.Int J Mol Sci. 2023 Oct 24;24(21):15518. doi: 10.3390/ijms242115518. Int J Mol Sci. 2023. PMID: 37958503 Free PMC article. Review.
-
Berberine Suppresses Influenza A Virus-Triggered Pyroptosis in Macrophages via Intervening in the mtROS-MAVS-NLRP3 Inflammasome Pathway.Viruses. 2025 Apr 7;17(4):539. doi: 10.3390/v17040539. Viruses. 2025. PMID: 40284982 Free PMC article.
-
A Review of Fibraurea tinctoria and Its Component, Berberine, as an Antidiabetic and Antioxidant.Molecules. 2023 Jan 29;28(3):1294. doi: 10.3390/molecules28031294. Molecules. 2023. PMID: 36770960 Free PMC article. Review.
References
-
- COVID‐19 Dashboard by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (JHU) . https://gisanddata.maps.arcgis.com/apps/opsdashboard/index.html#/bda7594.... Accessed December 1, 2020
-
- Dr. Fauci: US may see ‘surge upon surge’ of virus this winter. https://www.clickondetroit.com/health/2020/11/29/dr‐fauci‐us‐may‐see‐sur.... Accessed December 1, 2020
-
- Lee N, Hui D, Wu A, et al. A major outbreak of severe acute respiratory syndrome in Hong Kong. N Engl J Med. 2003;348(20):1986‐1994. - PubMed
-
- Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367(19):1814‐1820. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

