Disrupting upstream translation in mRNAs is associated with human disease
- PMID: 33750777
- PMCID: PMC7943595
- DOI: 10.1038/s41467-021-21812-1
Disrupting upstream translation in mRNAs is associated with human disease
Abstract
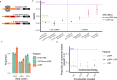
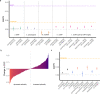
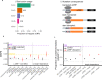
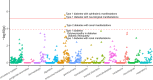
Ribosome-profiling has uncovered pervasive translation in non-canonical open reading frames, however the biological significance of this phenomenon remains unclear. Using genetic variation from 71,702 human genomes, we assess patterns of selection in translated upstream open reading frames (uORFs) in 5'UTRs. We show that uORF variants introducing new stop codons, or strengthening existing stop codons, are under strong negative selection comparable to protein-coding missense variants. Using these variants, we map and validate gene-disease associations in two independent biobanks containing exome sequencing from 10,900 and 32,268 individuals, respectively, and elucidate their impact on protein expression in human cells. Our results suggest translation disrupting mechanisms relating uORF variation to reduced protein expression, and demonstrate that translation at uORFs is genetically constrained in 50% of human genes.
Conflict of interest statement
A.B. is employed by Regeneron Pharmaceuticals. L.R.G. is employed by Janssen Research and Development. The rest of the authors declare no competing interests.
Figures

References
-
- uAUG and uORFs in human and rodent 5′untranslated mRNAs. Gene349, 97–105 (2005). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

