Critical role of the BAF chromatin remodeling complex during murine neural crest development
- PMID: 33750945
- PMCID: PMC8016319
- DOI: 10.1371/journal.pgen.1009446
Critical role of the BAF chromatin remodeling complex during murine neural crest development
Abstract
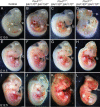
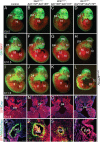
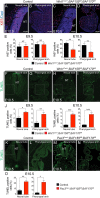
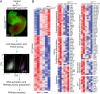
The BAF complex plays an important role in the development of a wide range of tissues by modulating gene expression programs at the chromatin level. However, its role in neural crest development has remained unclear. To determine the role of the BAF complex, we deleted BAF155/BAF170, the core subunits required for the assembly, stability, and functions of the BAF complex in neural crest cells (NCCs). Neural crest-specific deletion of BAF155/BAF170 leads to embryonic lethality due to a wide range of developmental defects including craniofacial, pharyngeal arch artery, and OFT defects. RNAseq and transcription factor enrichment analysis revealed that the BAF complex modulates the expression of multiple signaling pathway genes including Hippo and Notch, essential for the migration, proliferation, and differentiation of the NCCs. Furthermore, we demonstrated that the BAF complex is essential for the Brg1-Yap-Tead-dependent transcription of target genes in NCCs. Together, our results demonstrate an important role of the BAF complex in modulating the gene regulatory network essential for neural crest development.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Feiner L, Webber AL, Brown CB, Lu MM, Jia L, Feinstein P, et al.. Targeted disruption of semaphorin 3C leads to persistent truncus arteriosus and aortic arch interruption. Development. 2001;128(16):3061–70. . - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

