Mutation of the imprinted gene OsEMF2a induces autonomous endosperm development and delayed cellularization in rice
- PMID: 33751094
- PMCID: PMC8136911
- DOI: 10.1093/plcell/koaa006
Mutation of the imprinted gene OsEMF2a induces autonomous endosperm development and delayed cellularization in rice
Abstract
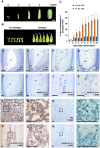
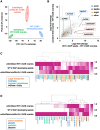
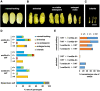
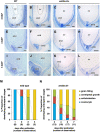
In angiosperms, endosperm development comprises a series of developmental transitions controlled by genetic and epigenetic mechanisms that are initiated after double fertilization. Polycomb repressive complex 2 (PRC2) is a key component of these mechanisms that mediate histone H3 lysine 27 trimethylation (H3K27me3); the action of PRC2 is well described in Arabidopsis thaliana but remains uncertain in cereals. In this study, we demonstrate that mutation of the rice (Oryza sativa) gene EMBRYONIC FLOWER2a (OsEMF2a), encoding a zinc-finger containing component of PRC2, causes an autonomous endosperm phenotype involving proliferation of the central cell nuclei with separate cytoplasmic domains, even in the absence of fertilization. Detailed cytological and transcriptomic analyses revealed that the autonomous endosperm can produce storage compounds, starch granules, and protein bodies specific to the endosperm. These events have not been reported in Arabidopsis. After fertilization, we observed an abnormally delayed developmental transition in the endosperm. Transcriptome and H3K27me3 ChIP-seq analyses using endosperm from the emf2a mutant identified downstream targets of PRC2. These included >100 transcription factor genes such as type-I MADS-box genes, which are likely required for endosperm development. Our results demonstrate that OsEMF2a-containing PRC2 controls endosperm developmental programs before and after fertilization.
© The Author(s) 2020. Published by Oxford University Press on behalf of American Society of Plant Biologists.
Figures


References
-
- Beauzamy L, Fourquin C, Dubrulle N, Boursiac Y, Boudaoud A, Ingram G (2016) Endosperm turgor pressure decreases during early Arabidopsis seed development. Development 143: 3295–3299 - PubMed

