miR-15a/16-1 deletion in activated B cells promotes plasma cell and mature B-cell neoplasms
- PMID: 33751108
- PMCID: PMC8033455
- DOI: 10.1182/blood.2020009088
miR-15a/16-1 deletion in activated B cells promotes plasma cell and mature B-cell neoplasms
Abstract
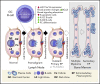
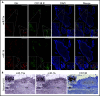
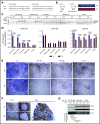
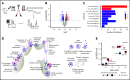
Chromosome 13q deletion [del(13q)], harboring the miR-15a/16-1 cluster, is one of the most common genetic alterations in mature B-cell malignancies, which originate from germinal center (GC) and post-GC B cells. Moreover, miR-15a/16 expression is frequently reduced in lymphoma and multiple myeloma (MM) cells without del(13q), suggesting important tumor-suppressor activity. However, the role of miR-15a/16-1 in B-cell activation and initiation of mature B-cell neoplasms remains to be determined. We show that conditional deletion of the miR-15a/16-1 cluster in murine GC B cells induces moderate but widespread molecular and functional changes including an increased number of GC B cells, percentage of dark zone B cells, and maturation into plasma cells. With time, this leads to development of mature B-cell neoplasms resembling human extramedullary plasmacytoma (EP) as well as follicular and diffuse large B-cell lymphomas. The indolent nature and lack of bone marrow involvement of EP in our murine model resembles human primary EP rather than MM that has progressed to extramedullary disease. We corroborate human primary EP having low levels of miR-15a/16 expression, with del(13q) being the most common genetic loss. Additionally, we show that, although the mutational profile of human EP is similar to MM, there are some exceptions such as the low frequency of hyperdiploidy in EP, which could account for different disease presentation. Taken together, our studies highlight the significant role of the miR-15a/16-1 cluster in the regulation of the GC reaction and its fundamental context-dependent tumor-suppression function in plasma cell and B-cell malignancies.
© 2021 by The American Society of Hematology.
Conflict of interest statement
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Figures


Similar articles
-
MiR-15a, miR-16-1 and miR-17-92 cluster expression are linked to poor prognosis in multiple myeloma.Leuk Res. 2012 Dec;36(12):1505-9. doi: 10.1016/j.leukres.2012.08.021. Epub 2012 Sep 7. Leuk Res. 2012. PMID: 22959509
-
Oncogenic Role of miR-15a-3p in 13q Amplicon-Driven Colorectal Adenoma-to-Carcinoma Progression.PLoS One. 2015 Jul 6;10(7):e0132495. doi: 10.1371/journal.pone.0132495. eCollection 2015. PLoS One. 2015. PMID: 26148070 Free PMC article.
-
MicroRNA-15a/16-1 cluster located at chromosome 13q14 is down-regulated but displays different expression pattern and prognostic significance in multiple myeloma.Oncotarget. 2015 Nov 10;6(35):38270-82. doi: 10.18632/oncotarget.5681. Oncotarget. 2015. PMID: 26516702 Free PMC article.
-
BCL2 and miR-15/16: from gene discovery to treatment.Cell Death Differ. 2018 Jan;25(1):21-26. doi: 10.1038/cdd.2017.159. Epub 2017 Oct 6. Cell Death Differ. 2018. PMID: 28984869 Free PMC article. Review.
-
Evaluation of MiR-15a and MiR-16-1 as prognostic biomarkers in chronic lymphocytic leukemia.Biomed Pharmacother. 2017 Aug;92:864-869. doi: 10.1016/j.biopha.2017.05.144. Epub 2017 Jun 6. Biomed Pharmacother. 2017. PMID: 28599250 Review.
Cited by
-
Opening new frontiers with catalytic nucleic acids in miRNA inhibition.Front Pharmacol. 2025 Jun 23;16:1604711. doi: 10.3389/fphar.2025.1604711. eCollection 2025. Front Pharmacol. 2025. PMID: 40626302 Free PMC article. Review.
-
Epigenetic Regulation of B Cell Memory Formation: A Poised Model for B Cell Epigenetic Reprograming.J Immunol Res. 2025 Jul 17;2025:9328523. doi: 10.1155/jimr/9328523. eCollection 2025. J Immunol Res. 2025. PMID: 40709286 Free PMC article. Review.
-
Laboratory Mice - A Driving Force in Immunopathology and Immunotherapy Studies of Human Multiple Myeloma.Front Immunol. 2021 Jun 2;12:667054. doi: 10.3389/fimmu.2021.667054. eCollection 2021. Front Immunol. 2021. PMID: 34149703 Free PMC article. Review.
-
BCL2 Protein Progressively Declines during Robust CLL Clonal Expansion: Potential Impact on Venetoclax Clinical Efficacy and Insights on Mechanism.Lymphatics. 2024 Jun;2(2):50-78. doi: 10.3390/lymphatics2020005. Epub 2024 Mar 28. Lymphatics. 2024. PMID: 39664277 Free PMC article.
-
The marine-derived compound TAG alleviates Parkinson's disease by restoring RUBCN-mediated lipid metabolism homeostasis.Acta Pharmacol Sin. 2024 Jul;45(7):1366-1380. doi: 10.1038/s41401-024-01259-y. Epub 2024 Mar 27. Acta Pharmacol Sin. 2024. PMID: 38538717 Free PMC article.
References
-
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281-297. - PubMed
-
- Bartel DP, Chen CZ. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet. 2004;5(5):396-400. - PubMed
-
- Selbach M, Schwanhäusser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455(7209):58-63. - PubMed
-
- Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834-838. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

