LncRNA-ZXF1 stabilizes P21 expression in endometrioid endometrial carcinoma by inhibiting ubiquitination-mediated degradation and regulating the miR-378a-3p/PCDHA3 axis
- PMID: 33751805
- PMCID: PMC8807357
- DOI: 10.1002/1878-0261.12940
LncRNA-ZXF1 stabilizes P21 expression in endometrioid endometrial carcinoma by inhibiting ubiquitination-mediated degradation and regulating the miR-378a-3p/PCDHA3 axis
Abstract
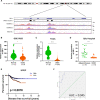
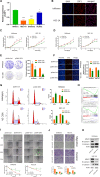
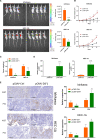
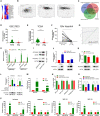
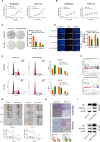
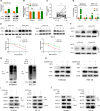
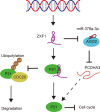
Long noncoding RNAs (lncRNAs) have a profound effect on biological processes in various malignancies. However, few studies have investigated their functions and specific mechanisms in endometrial cancer. In this study, we focused on the role and mechanism of lncRNA-ZXF1 in endometrial cancer. Bioinformatics and in vitro and in vivo experiments were used to explore the expression and function of lncRNA-ZXF1. We found that lncRNA-ZXF1 altered the migration and invasion of endometrioid endometrial cancer (EEC) cells. Furthermore, our results suggest that lncRNA-ZXF1 regulates EEC cell proliferation. This regulation may be achieved by the lncRNA-ZXF1-mediated alteration in the expression of P21 through two mechanisms. One is that lncRNA-ZXF1 functions as a molecular sponge of miR-378a-3p to regulate PCDHA3 expression and then modulate the expression of P21. The other is that lncRNA-ZXF1 inhibits CDC20-mediated degradation of ubiquitination by directly binding to P21. To the best of our knowledge, this study is the first to explore lncRNA-ZXF1 functioning as a tumor-suppressing lncRNA in EEC. LncRNA-ZXF1 may become therapeutic, diagnostic, and prognostic indicator in the future.
Keywords: P21; cell cycle; endometrioid endometrial cancer; lncRNA-ZXF1; ubiquitination.
© 2021 The Authors. Molecular Oncology published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Enhanced expression of long non-coding RNA ZXF1 promoted the invasion and metastasis in lung adenocarcinoma.Biomed Pharmacother. 2014 May;68(4):401-7. doi: 10.1016/j.biopha.2014.03.001. Epub 2014 Mar 19. Biomed Pharmacother. 2014. PMID: 24721325
-
Disrupting MALAT1/miR-200c sponge decreases invasion and migration in endometrioid endometrial carcinoma.Cancer Lett. 2016 Dec 1;383(1):28-40. doi: 10.1016/j.canlet.2016.09.019. Epub 2016 Sep 28. Cancer Lett. 2016. PMID: 27693631
-
Enhanced expression of lncRNA ZXF1 promotes cisplatin resistance in lung cancer cell via MAPK axis.Exp Mol Pathol. 2020 Oct;116:104484. doi: 10.1016/j.yexmp.2020.104484. Epub 2020 Jun 10. Exp Mol Pathol. 2020. PMID: 32533982
-
LINC00958 promotes endometrial cancer cell proliferation and metastasis by regulating the miR-145-3p/TCF4 axis.J Gene Med. 2021 Jul;23(7):e3345. doi: 10.1002/jgm.3345. Epub 2021 May 11. J Gene Med. 2021. PMID: 33885186
-
LncRNA NEAT1 promotes endometrial cancer cell proliferation, migration and invasion by regulating the miR-144-3p/EZH2 axis.Radiol Oncol. 2019 Nov 20;53(4):434-442. doi: 10.2478/raon-2019-0051. Radiol Oncol. 2019. PMID: 31747378 Free PMC article.
Cited by
-
lncRNA ACTA2-AS1 inhibits malignant phenotypes of gastric cancer cells.Open Med (Wars). 2022 Feb 15;17(1):266-279. doi: 10.1515/med-2021-0406. eCollection 2022. Open Med (Wars). 2022. Retraction in: Open Med (Wars). 2023 Dec 31;18(1):20230892. doi: 10.1515/med-2023-0892. PMID: 35274046 Free PMC article. Retracted.
-
The Promotive and Inhibitory Role of Long Non-Coding RNAs in Endometrial Cancer Course-A Review.Cancers (Basel). 2024 Jun 3;16(11):2125. doi: 10.3390/cancers16112125. Cancers (Basel). 2024. PMID: 38893244 Free PMC article. Review.
-
LncRNA linc01194 promotes the progress of endometrial carcinoma by up-regulating SOX2 through binding to IGF2BP1.J Gynecol Oncol. 2024 Mar;35(2):e21. doi: 10.3802/jgo.2024.35.e21. Epub 2023 Nov 21. J Gynecol Oncol. 2024. PMID: 38072399 Free PMC article.
-
The E3 Ligases in Cervical Cancer and Endometrial Cancer.Cancers (Basel). 2022 Oct 30;14(21):5354. doi: 10.3390/cancers14215354. Cancers (Basel). 2022. PMID: 36358773 Free PMC article. Review.
-
N4-acetylcytidine modification of LncRNA GFOD1-AS1 promotes high glucose-induced dysfunction in human dermal microvascular endothelial cells through stabilization of DNMT1 protein.Funct Integr Genomics. 2025 May 24;25(1):107. doi: 10.1007/s10142-025-01617-x. Funct Integr Genomics. 2025. PMID: 40411601 Free PMC article.
References
-
- Rebecca LS, Kimberly DM & Ahmedin J (2020) Cancer statistics, 2020. CA Cancer J Clin 70, 7–30. - PubMed
-
- Rebecca LS, Kimberly DM & Ahmedin J (2019) Cancer statistics, 2019. CA Cancer J Clin 69, 7–34. - PubMed
-
- Kimberly DM, Leticia N, Angela BM, Julia HRK, Robin Y, Catherine MA, Ahmedin J, Joan LK & Rebecca LS (2019) Cancer treatment and survivorship statistics, 2019. CA Cancer J Clin 65, 363–385.
-
- Lheureux S & Oza AM (2016) Endometrial cancer‐targeted therapies myth or reality Review of current targeted treatments. Eur J Cancer 59, 99–108. - PubMed
-
- Brooks RA, Fleming GF, Lastra RR, Lee NK, Moroney JW, Son CH, Tatebe K & Veneris JL (2019) Current recommendations and recent progress in endometrial cancer. CA Cancer J Clin 69, 258–279. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

