Copy number variation and neuropsychiatric illness
- PMID: 33752146
- PMCID: PMC8219524
- DOI: 10.1016/j.gde.2021.02.014
Copy number variation and neuropsychiatric illness
Abstract
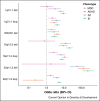
Copy number variants (CNVs) at specific loci have been identified as important risk factors for several neuropsychiatric disorders, such as schizophrenia, autism spectrum disorder, intellectual disability (ID) and depression. These CNVs are individually rare (<0.5% frequency), have high effect sizes, and show pleiotropic effects for multiple neuropsychiatric disorders, which implies a shared aetiology. Neuropsychiatric CNVs are also associated with cognitive impairment and other medical morbidities, such as heart defects and obesity. As most neuropsychiatric CNVs are multigenic, it has been challenging to map their effects onto specific biological processes, although gene-set analyses have implicated genes related to the synapse and chromatin regulation. However, future whole-genome sequencing studies have potential for identifying novel single-gene CNV associations, which could provide insights into the pathophysiology underlying neuropsychiatric disorders.
Copyright © 2021 The Authors. Published by Elsevier Ltd.. All rights reserved.
Figures

References
-
- Sebat J., Lakshmi B., Troge J., Alexander J., Young J., Lundin P., Månér S., Massa H., Walker M., Chi M. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. - PubMed
-
- Collins R.L., Brand H., Karczewski K.J., Zhao X., Alföldi J., Francioli L.C., Khera A.V., Lowther C., Gauthier L.D., Wang H. A structural variation reference for medical and population genetics. Nature. 2020;581:444–451. - PMC - PubMed
-
This landmark paper presents the first reference map of structural variants (SVs) called high-coverage whole genome sequencing data. The paper evaluates the frequencies and distribution of deletions, duplications, multiallelic CNVs, insertions, inversions, translocations and complex SVs across diverse populations using data from 14 891 individuals. The authors report that SVs contribute between 25–59% of all protein truncating events per genome, and that the patterns of selection which operate against SVs are similar to those observed for single nucleotide variants. The SVs reported in this paper are freely available through the gnomAD browser (https://gnomad.broadinstitute.org/). This resource will greatly aid the interpretation of SVs discovered in future whole-genome sequencing studies of neuropsychiatric disorders.
-
- Murphy K.C., Jones L.A., Owen M.J. High rates of schizophrenia in adults with velo-cardio-facial syndrome. Arch Gen Psychiatry. 1999;56:940–945. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

