GRAS-domain transcription factor PAT1 regulates jasmonic acid biosynthesis in grape cold stress response
- PMID: 33752238
- PMCID: PMC8260143
- DOI: 10.1093/plphys/kiab142
GRAS-domain transcription factor PAT1 regulates jasmonic acid biosynthesis in grape cold stress response
Abstract
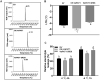
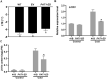
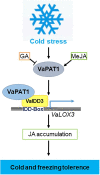
Cultivated grapevine (Vitis) is a highly valued horticultural crop, and cold stress affects its growth and productivity. Wild Amur grape (Vitis amurensis) PAT1 (Phytochrome A signal transduction 1, VaPAT1) is induced by low temperature, and ectopic expression of VaPAT1 enhances cold tolerance in Arabidopsis (Arabidopsis thaliana). However, little is known about the molecular mechanism of VaPAT1 during the cold stress response in grapevine. Here, we confirmed the overexpression of VaPAT1 in transformed grape calli enhanced cold tolerance. Yeast two-hybrid and bimolecular fluorescence complementation assays highlighted an interaction between VaPAT1 with INDETERMINATE-DOMAIN 3 (VaIDD3). A role of VaIDD3 in cold tolerance was also indicated. Transcriptome analysis revealed VaPAT1 and VaIDD3 overexpression and cold treatment coordinately modulate the expression of stress-related genes including lipoxygenase 3 (LOX3), a gene encoding a key jasmonate biosynthesis enzyme. Co-expression network analysis indicated LOX3 might be a downstream target of VaPAT1. Both electrophoretic mobility shift and dual luciferase reporter assays showed the VaPAT1-IDD3 complex binds to the IDD-box (AGACAAA) in the VaLOX3 promoter to activate its expression. Overexpression of both VaPAT1 and VaIDD3 increased the transcription of VaLOX3 and JA levels in transgenic grape calli. Conversely, VaPAT1-SRDX (dominant repression) and CRISPR/Cas9-mediated mutagenesis of PAT1-ED causing the loss of the C-terminus in grape calli dramatically prohibited the accumulation of VaLOX3 and JA levels during cold treatment. Together, these findings point to a pivotal role of VaPAT1 in the cold stress response in grape by regulating JA biosynthesis.
© American Society of Plant Biologists 2021. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures





References
-
- Ba L, Jianfei K, Chen J, Lu W (2016) MaJAZ1 attenuates the MaLBD5-mediated transcriptional activation of jasmonate biosynthesis gene MaAOC2 in regulating cold tolerance of banana fruit. J Agric Food Chem 64:738–745 - PubMed
-
- Bolle C (2004) The role of GRAS proteins in plant signal transduction and development. Planta 218:683–692 - PubMed
-
- Böttcher C, Burbidge CA, di Rienzo V, Boss PK, Davies C (2015) Jasmonic acid-isoleucine formation in grapevine (Vitis vinifera L.) by two enzymes with distinct transcription profiles. J Integr Plant Biol 57:618–627 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

