Reduce, Retain, Recycle: Mechanisms for Promoting Histone Protein Degradation versus Stability and Retention
- PMID: 33753462
- PMCID: PMC8316141
- DOI: 10.1128/MCB.00007-21
Reduce, Retain, Recycle: Mechanisms for Promoting Histone Protein Degradation versus Stability and Retention
Abstract
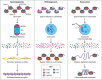
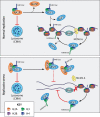
The eukaryotic genome is packaged into chromatin. The nucleosome, the basic unit of chromatin, is composed of DNA coiled around a histone octamer. Histones are among the longest-lived protein species in mammalian cells due to their thermodynamic stability and their associations with DNA and histone chaperones. Histone metabolism plays an integral role in homeostasis. While histones are largely stable, the degradation of histone proteins is necessary under specific conditions. Here, we review the physiological and cellular contexts that promote histone degradation. We describe specific known mechanisms that drive histone proteolysis. Finally, we discuss the importance of histone degradation and regulation of histone supply for organismal and cellular fitness.
Keywords: DNA replication; chaperone; chromatin; histone; nucleosome; nucleus; posttranslational modification; transcription.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
