Genome-wide identification of MIKC-type genes related to stamen and gynoecium development in Liriodendron
- PMID: 33753780
- PMCID: PMC7985208
- DOI: 10.1038/s41598-021-85927-7
Genome-wide identification of MIKC-type genes related to stamen and gynoecium development in Liriodendron
Abstract
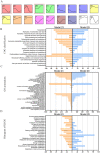
The organogenesis and development of reproductive organs, i.e., stamen and gynoecium, are important floral characteristics that are closely related to pollinators and reproductive fitness. As a genus from Magnoliaceae, Liriodendron has only two relict species: L. chinense and L. tulipifera. Despite the similar flower shapes of these species, their natural seed-setting rates differ significantly, implying interspecies difference in floral organogenesis and development. MADS-box genes, which participate in floral organogenesis and development, remain unexplored in Liriodendron. Here, to explore the interspecies difference in floral organogenesis and development and identify MADS-box genes in Liriodendron, we examined the stamen and gynoecium primordia of the two Liriodendron species by scanning electron microscopy combined with paraffin sectioning, and then collected two types of primordia for RNA-seq. A total of 12 libraries were constructed and 42,268 genes were identified, including 35,269 reference genes and 6,999 new genes. Monoterpenoid biosynthesis was enriched in L. tulipifera. Genome-wide analysis of 32 MADS-box genes was conducted, including phylogenetic trees, exon/intron structures, and conserved motif distributions. Twenty-six genes were anchored on 17 scaffolds, and six new genes had no location information. The expression profiles of MIKC-type genes via RT-qPCR acrossing six stamen and gynoecium developmental stages indicates that the PI-like, AG/STK-like, SEP-like, and SVP-like genes may contribute to the species-specific differentiation of the organogenesis and development of reproductive organs in Liriodendron. Our findings laid the groundwork for the future exploration of the mechanism underlying on the interspecific differences in reproductive organ development and fitness in Liriodendron.
Conflict of interest statement
The authors declare no competing interests.
Figures









References
-
- Li HG, Chen L, Liang CY, Huang MR. A case study on provenance testing of tulip tree (Liriodendron spp) Chin. For. Sci. Technol. 2005;19(5):6–13. doi: 10.3969/j.issn.1000-8101.2005.05.005. - DOI
-
- Xu F, Rudall PJ. Comparative floral anatomy and ontogeny in Magnoliaceae. Plant Syst. Evol. 2006;258:1–15. doi: 10.1007/s00606-005-0361-1. - DOI
-
- Wang S, Xie Y. Red List of Endangered Plants in China. Beijing; 2004.
-
- Fan RW, Ye JG, Yin ZF, Gao HG, You LX. Studies on seed development and embryogenesis in Liriodendron chinense (Hemsl) Sary. Acta Bot. Sin. 1992;34(6):437–442.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

