Discovery and resistance mechanism of a selective CDK12 degrader
- PMID: 33753926
- PMCID: PMC8590456
- DOI: 10.1038/s41589-021-00765-y
Discovery and resistance mechanism of a selective CDK12 degrader
Abstract
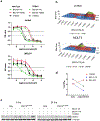
Cyclin-dependent kinase 12 (CDK12) is an emerging therapeutic target due to its role in regulating transcription of DNA-damage response (DDR) genes. However, development of selective small molecules targeting CDK12 has been challenging due to the high degree of homology between kinase domains of CDK12 and other transcriptional CDKs, most notably CDK13. In the present study, we report the rational design and characterization of a CDK12-specific degrader, BSJ-4-116. BSJ-4-116 selectively degraded CDK12 as assessed through quantitative proteomics. Selective degradation of CDK12 resulted in premature cleavage and poly(adenylation) of DDR genes. Moreover, BSJ-4-116 exhibited potent antiproliferative effects, alone and in combination with the poly(ADP-ribose) polymerase inhibitor olaparib, as well as when used as a single agent against cell lines resistant to covalent CDK12 inhibitors. Two point mutations in CDK12 were identified that confer resistance to BSJ-4-116, demonstrating a potential mechanism that tumor cells can use to evade bivalent degrader molecules.
Figures






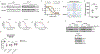
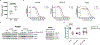
References
METHODS ONLY REFERENCES
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

