Screening and bioinformatical analysis of differentially expressed genes in nasopharyngeal carcinoma
- PMID: 33753985
- PMCID: PMC7974527
- DOI: 10.7150/jca.48979
Screening and bioinformatical analysis of differentially expressed genes in nasopharyngeal carcinoma
Abstract
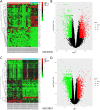
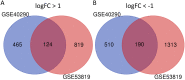
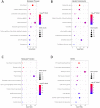
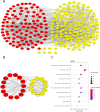
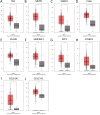
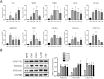
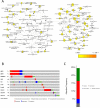
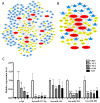
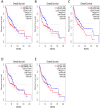
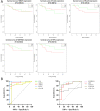
Objective: To identify differentially expressed genes via bioinformatical analysis for nasopharyngeal carcinoma (NPC) and explore potential biomarkers for NPC. Methods: We downloaded the NPC gene expression datasets (GSE40290, GSE53819) and obtained differentially expressed genes (DEGs) via GEO2R. Functional analysis of DEGs was performed by Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis. In order to explore the interaction of DEGs and screen the core genes, we established protein-protein interaction (PPI) network. Then the expression level, prognostic and diagnostic analysis of the core genes in NPC were performed to reveal their potential effects on NPC. Furthermore, we obtained the transcription factors (TF) and microRNAs of core genes to construct the coregulatory network. Results: We obtained 124 up-regulated genes and 190 down-regulated genes in total. These genes were found to be related to signal transduction, extracellular matrix organization and cell adhesion based on GO analysis. KEGG analysis revealed that the NF-kappa B (NF-κB) signaling pathway, pathways in cancer were mainly enriched signaling pathways. 25 core genes were obtained by constructing PPI network. Then the high expression of 10 core genes in NPC were verified via GEPIA, Oncomine databases and laboratory experiments. The TF-microRNA coregulatory network of the 10 core genes was built. Survival and diagnostic analysis indicated that SPP1 had negative influence on the prognosis of NPC patients based on two datasets and nine up-regulated core genes (FN1, MMP1, MMP3, PLAU, PLAUR, SERPINE1, SPP1, COL8A1, COL10A1) might be diagnostic markers for NPC. Conclusions: Core genes of NPC were screened out by bioinformatical analysis in the present study and these genes may serve as prognostic and diagnostic biomarkers for NPC.
Keywords: bioinformatical analysis; core genes; microarray; nasopharyngeal carcinoma; prognosis.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures

References
-
- Chua MLK, Wee JTS, Hui EP. et al. Nasopharyngeal carcinoma. Lancet. 2016;387:1012–1024. - PubMed
-
- Ng WT, Wong ECY, Lee VHF. et al. Head and neck cancer in Hong Kong. Jpn J Clin Oncol. 2018;48:13–21. - PubMed
-
- Roy Chattopadhyay N, Das P, Chatterjee K. et al. Higher incidence of nasopharyngeal carcinoma in some regions in the world confers for interplay between genetic factors and external stimuli. Drug Discov Ther. 2017;11:170–180. - PubMed
-
- Zhang J, Jia L, Tsang CM. et al. EBV infection and glucose metabolism in nasopharyngeal carcinoma. Adv Exp Med Biol. 2017;1018:75–90. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

