Candida albicans disorder is associated with gastric carcinogenesis
- PMID: 33754037
- PMCID: PMC7978306
- DOI: 10.7150/thno.55209
Candida albicans disorder is associated with gastric carcinogenesis
Abstract
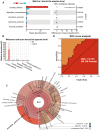
Background: Bacterial infection is associated with gastric carcinogenesis. However, the relationship between nonbacterial components and gastric cancer (GC) has not been fully explored. We aimed to characterize the fungal microbiome in GC. Methods: We performed ITS rDNA gene analysis in cancer lesions and adjacent noncancerous tissues of 45 GC cases from Shenyang, China. Obtaining the OTUs and combining effective grouping, we carried out species identifications, alpha and beta diversity analyses, and FUNGuild functional annotation. Moreover, differences were compared and tested between groups to better investigate the composition and ecology of fungi associated with GC and find fungal indicators. Results: We observed significant gastric fungal imbalance in GC. Principal component analysis revealed separate clusters for the GC and control groups, and Venn diagram analysis indicated that the GC group showed a lower OTU abundance than the control. At the genus level, the abundances of 15 fungal biomarkers distinguished the GC group from the control, of which Candida (p = 0.000246) and Alternaria (p = 0.00341) were enriched in GC, while Saitozyma (p = 0.002324) and Thermomyces (p = 0.009158) were decreased. Combining the results of Welch's t test and Wilcoxon rank sum test, Candida albicans (C. albicans) was significantly elevated in GC. The species richness Krona pie chart further revealed that C. albicans occupied 22% and classified GC from the control with an area under the receiver operating curve (AUC) of 0.743. Random forest analysis also confirmed that C. albicans could serve as a biomarker with a certain degree of accuracy. Moreover, compared with that of the control, the alpha diversity index was significantly reduced in the GC group. The Jaccard distance index and the Bray abundance index of the PCoA clarified separate clusters between the GC and control groups at the species level (p = 0.00051). Adonis (PERMANOVA) analysis and ANOVA showed that there were significant differences in fungal structure among groups (p = 0.001). Finally, FUNGuild functional classification predicted that saprotrophs were the most abundant taxa in the GC group. Conclusions: This study revealed GC-associated mycobiome imbalance characterized by an altered fungal composition and ecology and demonstrated that C. albicans can be a fungal biomarker for GC. With the significant increase of C. albicans in GC, the abundance of Fusicolla acetilerea, Arcopilus aureus, Fusicolla aquaeductuum were increased, while Candida glabrata, Aspergillus montevidensis, Saitozyma podzolica and Penicillium arenicola were obviously decreased. In addition, C. albicans may mediate GC by reducing the diversity and richness of fungi in the stomach, contributing to the pathogenesis of GC.
Keywords: Candida albicans; Gastric cancer; biomarker; fungal imbalance; mycobiome.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures





References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

