Deciphering the autophagy regulatory network via single-cell transcriptome analysis reveals a requirement for autophagy homeostasis in spermatogenesis
- PMID: 33754041
- PMCID: PMC7978313
- DOI: 10.7150/thno.55645
Deciphering the autophagy regulatory network via single-cell transcriptome analysis reveals a requirement for autophagy homeostasis in spermatogenesis
Abstract
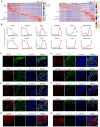
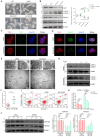
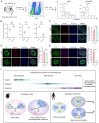
Background: Autophagy has been implicated as a crucial component in spermatogenesis, and autophagy dysfunction can lead to reproductive disorders in animal models, including yeast, C. elegans and mice. However, the sophisticated transcriptional networks of autophagic genes throughout human spermatogenesis and their biological significance remain largely uncharacterized. Methods: We profiled the transcriptional signatures of autophagy-related genes during human spermatogenesis by assessing specimens from nine fertile controls (including two normal persons and seven obstructive azoospermia (OA) patients) and one nonobstructive azoospermia (NOA) patient using single-cell RNA sequencing (scRNA-seq) analysis. Dysregulation of autophagy was confirmed in two additional NOA patients by immunofluorescence staining. Gene knockdown was used to identify the role of Cst3 in autophagy during spermatogenesis. Results: Our data uncovered a unique, global stage-specific enrichment of autophagy-related genes. Human-mouse comparison analysis revealed that the stage-specific expression pattern of autophagy-related genes was highly conserved in mammals. More importantly, dysregulation of some clusters of autophagy-related genes was observed in NOA patients, suggesting the association of autophagy with male infertility. Cst3, a human-mouse conserved and autophagy-related gene that is actively expressed in spermatogonia and early spermatocytes, was found to regulate spermatogonial stem cell (SSC) maintenance and subsequent male germ cell development. Knockdown of Cst3 increased autophagic activity in mouse SSCs and subsequently suppressed the transcription of SSC core factors such as Oct4, Id1, and Nanos3, which could be efficiently rescued by manipulating autophagic activity. Conclusions: Our study provides comprehensive insights into the global transcriptional signatures of autophagy-related genes and confirms the importance of autophagy homeostasis in SSC maintenance and normal spermatogenesis, opening new avenues for further dissecting the significance of the autophagy regulatory network in spermatogenesis as well as male infertility.
Keywords: autophagy; male infertility.; meiosis; single-cell RNA sequencing; spermatogenesis; spermatogonial stem cells.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures


References
-
- Zuccarello D, Ferlin A, Cazzadore C, Pepe A, Garolla A, Moretti A. et al. Mutations in dynein genes in patients affected by isolated non-syndromic asthenozoospermia. Hum Reprod. 2008;23:1957–62. - PubMed
-
- Heidary Z, Saliminejad K, Zaki-Dizaji M, Khorram Khorshid HR. Genetic aspects of idiopathic asthenozoospermia as a cause of male infertility. Hum Fertil (Camb) 2018. pp. 1–10. - PubMed
-
- Li L, Sha YW, Su ZY, Mei LB, Ji ZY, Zhang Q. et al. A novel mutation in HAUS7 results in severe oligozoospermia in two brothers. Gene. 2018;639:106–10. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

