Recent advances of Cas12a applications in bacteria
- PMID: 33754170
- PMCID: PMC8053165
- DOI: 10.1007/s00253-021-11243-9
Recent advances of Cas12a applications in bacteria
Abstract
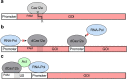
Clustered regularly interspaced short palindromic repeats (CRISPR)-mediated genome engineering and related technologies have revolutionized biotechnology over the last decade by enhancing the efficiency of sophisticated biological systems. Cas12a (Cpf1) is an RNA-guided endonuclease associated to the CRISPR adaptive immune system found in many prokaryotes. Contrary to its more prominent counterpart Cas9, Cas12a recognizes A/T rich DNA sequences and is able to process its corresponding guide RNA directly, rendering it a versatile tool for multiplex genome editing efforts and other applications in biotechnology. While Cas12a has been extensively used in eukaryotic cell systems, microbial applications are still limited. In this review, we highlight the mechanistic and functional differences between Cas12a and Cas9 and focus on recent advances of applications using Cas12a in bacterial hosts. Furthermore, we discuss advantages as well as current challenges and give a future outlook for this promising alternative CRISPR-Cas system for bacterial genome editing and beyond. KEY POINTS: • Cas12a is a powerful tool for genome engineering and transcriptional perturbation • Cas12a causes less toxic side effects in bacteria than Cas9 • Self-processing of crRNA arrays facilitates multiplexing approaches.
Keywords: CRISPR-Cas12a; Genome editing; Multiplex gene regulation; Transcriptional perturbation.
Conflict of interest statement
The authors declare no competing interests.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

