Association of sigma-1 receptor with dopamine transporter attenuates the binding of methamphetamine via distinct helix-helix interactions
- PMID: 33754484
- PMCID: PMC8113090
- DOI: 10.1111/cbdd.13841
Association of sigma-1 receptor with dopamine transporter attenuates the binding of methamphetamine via distinct helix-helix interactions
Abstract
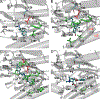
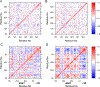
Dopamine transporter (DAT) and sigma-1 receptor (σ1R) are potential therapeutic targets to reduce the psychostimulant effects induced by methamphetamine (METH). Interaction of σ1R with DAT could modulate the binding of METH, but the molecular basis of the association of the two transmembrane proteins and how their interactions mediate the binding of METH to DAT or σ1R remain unclear. Here, we characterize the protein-ligand and protein-protein interactions at a molecular level by various theoretical approaches. The present results show that METH adopts a different binding pose in the binding pocket of σ1R and is more likely to act as an agonist. The relatively lower binding affinity of METH to σ1R supports the role of antagonists as inhibitors that protect against METH-induced effects. We demonstrate that σ1R could bind to Drosophila melanogaster DAT (dDAT) through interactions with either the transmembrane helix α12 or α5 of dDAT. Our results showed that the truncated σ1R displays stronger association with dDAT than the full-length σ1R. Although different helix-helix interactions between σ1R and dDAT lead to distinct effects on the dynamics of individual protein, both associations attenuate the binding affinity of METH to dDAT, particularly in the interactions with the helix α5 of dDAT. Together, the present study provides the first computational investigation on the molecular mechanism of coupling METH binding and the association of σ1R with dDAT.
Keywords: association; binding affinity; dopamine transporter; methamphetamine; sigma-1 receptor.
© 2021 John Wiley & Sons A/S.
Conflict of interest statement
CONFLICT OF INTEREST
The authors declare no competing financial interest.
Figures



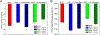

References
-
- Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, & Lindahl E (2015). Gromacs: High Performance Molecular Simulations through Multi-Level Parallelism from Laptops to Supercomputers. SoftwareX, 1–2, 19–25. doi:10.1016/j.softx.2015.06.001 - DOI
-
- Amadei A, Ceruso MA, & Di Nola A (1999). On the Convergence of the Conformational Coordinates Basis Set Obtained by the Essential Dynamics Analysis of Proteins’ Molecular Dynamics Simulations. Proteins: Structure, Function, and Genetics, 36, 419–424. doi:10.1002/(sici)1097-0134(19990901)36:4<419::aid-prot5>3.0.co;2-u - DOI - PubMed
-
- Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, & Haak JR (1984). Molecular Dynamics with Coupling to an External Bath. The Journal of Chemical Physics, 81, 3684–3690. doi:10.1063/1.448118 - DOI
-
- Berendsen HJC, van der Spoel D, & van Drunen R (1995). Gromacs: A Message-Passing Parallel Molecular Dynamics Implementation. Computer Physics Communications, 91, 43–56. doi:10.1016/0010-4655(95)00042-e - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

