Model-based analysis and forecast of sleep-wake regulatory dynamics: Tools and applications to data
- PMID: 33754773
- PMCID: PMC7837756
- DOI: 10.1063/5.0024024
Model-based analysis and forecast of sleep-wake regulatory dynamics: Tools and applications to data
Abstract
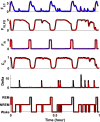
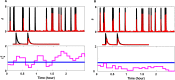
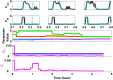
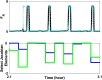
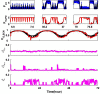
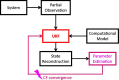
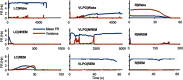
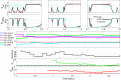
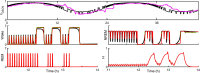
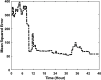
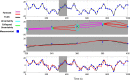
Extensive clinical and experimental evidence links sleep-wake regulation and state of vigilance (SOV) to neurological disorders including schizophrenia and epilepsy. To understand the bidirectional coupling between disease severity and sleep disturbances, we need to investigate the underlying neurophysiological interactions of the sleep-wake regulatory system (SWRS) in normal and pathological brains. We utilized unscented Kalman filter based data assimilation (DA) and physiologically based mathematical models of a sleep-wake regulatory network synchronized with experimental measurements to reconstruct and predict the state of SWRS in chronically implanted animals. Critical to applying this technique to real biological systems is the need to estimate the underlying model parameters. We have developed an estimation method capable of simultaneously fitting and tracking multiple model parameters to optimize the reconstructed system state. We add to this fixed-lag smoothing to improve reconstruction of random input to the system and those that have a delayed effect on the observed dynamics. To demonstrate application of our DA framework, we have experimentally recorded brain activity from freely behaving rodents and classified discrete SOV continuously for many-day long recordings. These discretized observations were then used as the "noisy observables" in the implemented framework to estimate time-dependent model parameters and then to forecast future state and state transitions from out-of-sample recordings.
Figures


References
-
- Datta S. and MacLean R. R., “Neurobiological mechanisms for the regulation of mammalian sleep-wake behavior: Reinterpretation of historical evidence and inclusion of contemporary cellular and molecular evidence,” Neurosci. Biobehav. Rev. 31, 775–824 (2007). 10.1016/j.neubiorev.2007.02.004 - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

