Antigenic and molecular characterization of low pathogenic avian influenza A(H9N2) viruses in sub-Saharan Africa from 2017 through 2019
- PMID: 33754959
- PMCID: PMC8057090
- DOI: 10.1080/22221751.2021.1908097
Antigenic and molecular characterization of low pathogenic avian influenza A(H9N2) viruses in sub-Saharan Africa from 2017 through 2019
Abstract
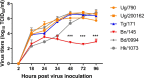
Sub-Saharan Africa was historically considered an animal influenza cold spot, with only sporadic highly pathogenic H5 outbreaks detected over the last 20 years. However, in 2017, low pathogenic avian influenza A(H9N2) viruses were detected in poultry in Sub-Saharan Africa. Molecular, phylogenetic, and antigenic characterization of isolates from Benin, Togo, and Uganda showed that they belonged to the G1 lineage. Isolates from Benin and Togo clustered with viruses previously described in Western Africa, whereas viruses from Uganda were genetically distant and clustered with viruses from the Middle East. Viruses from Benin exhibited decreased cross-reactivity with those from Togo and Uganda, suggesting antigenic drift associated with reduced replication in Calu-3 cells. The viruses exhibited mammalian adaptation markers similar to those of the human strain A/Senegal/0243/2019 (H9N2). Therefore, viral genetic and antigenic surveillance in Africa is of paramount importance to detect further evolution or emergence of new zoonotic strains.
Keywords: Africa; Influenza virus; antigenic cartography; one health; phylogeny.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures


Similar articles
-
Molecular and Antigenic Characterization of Avian H9N2 Viruses in Southern China.Microbiol Spectr. 2022 Feb 23;10(1):e0082221. doi: 10.1128/spectrum.00822-21. Epub 2022 Jan 12. Microbiol Spectr. 2022. PMID: 35019707 Free PMC article.
-
Immune Escape Variants of H9N2 Influenza Viruses Containing Deletions at the Hemagglutinin Receptor Binding Site Retain Fitness In Vivo and Display Enhanced Zoonotic Characteristics.J Virol. 2017 Jun 26;91(14):e00218-17. doi: 10.1128/JVI.00218-17. Print 2017 Jul 15. J Virol. 2017. PMID: 28468875 Free PMC article.
-
Genetic and antigenic characterization of avian influenza H9N2 viruses during 2016 in Iraq.Open Vet J. 2019 Jul;9(2):164-171. doi: 10.4314/ovj.v9i2.12. Epub 2019 Jun 12. Open Vet J. 2019. PMID: 31360657 Free PMC article.
-
H9N2 influenza virus in China: a cause of concern.Protein Cell. 2015 Jan;6(1):18-25. doi: 10.1007/s13238-014-0111-7. Epub 2014 Nov 11. Protein Cell. 2015. PMID: 25384439 Free PMC article. Review.
-
Sporadic occurrence of H9N2 avian influenza infections in human in Anhui province, eastern China: A notable problem.Microb Pathog. 2020 Mar;140:103940. doi: 10.1016/j.micpath.2019.103940. Epub 2019 Dec 18. Microb Pathog. 2020. PMID: 31863839
Cited by
-
Airborne Transmission of Avian Origin H9N2 Influenza A Viruses in Mammals.Viruses. 2021 Sep 24;13(10):1919. doi: 10.3390/v13101919. Viruses. 2021. PMID: 34696349 Free PMC article. Review.
-
Genetic insights of H9N2 avian influenza viruses circulating in Mali and phylogeographic patterns in Northern and Western Africa.Virus Evol. 2024 Feb 19;10(1):veae011. doi: 10.1093/ve/veae011. eCollection 2024. Virus Evol. 2024. PMID: 38435712 Free PMC article.
-
Genetic Evolution of Avian Influenza A (H9N2) Viruses Isolated from Domestic Poultry in Uganda Reveals Evidence of Mammalian Host Adaptation, Increased Virulence and Reduced Sensitivity to Baloxavir.Viruses. 2022 Sep 18;14(9):2074. doi: 10.3390/v14092074. Viruses. 2022. PMID: 36146881 Free PMC article.
-
Risk distribution of human infections with avian influenza A (H5N1, H5N6, H9N2 and H7N9) viruses in China.Front Public Health. 2024 Oct 24;12:1448974. doi: 10.3389/fpubh.2024.1448974. eCollection 2024. Front Public Health. 2024. PMID: 39512713 Free PMC article.
-
Surveillance and Phylogenetic Characterisation of Avian Influenza Viruses Isolated from Wild Waterfowl in Zambia in 2015, 2020, and 2021.Transbound Emerg Dis. 2023 Mar 1;2023:4606850. doi: 10.1155/2023/4606850. eCollection 2023. Transbound Emerg Dis. 2023. PMID: 40303760 Free PMC article.
References
-
- Peiris M, Yuen KY, Leung CW, et al. . Human infection with influenza H9N2. Lancet Lond Engl. 1999;354:916–917. - PubMed
-
- Cui L, Liu D, Shi W, et al. . Dynamic reassortments and genetic heterogeneity of the human-infecting influenza A (H7N9) virus. Nat Commun. 2014;5:1–9. - PubMed
-
- Liu D, Shi W, Gao GF.. Poultry carrying H9N2 act as incubators for novel human avian influenza viruses. Lancet Lond Engl. 2014;383:869. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
