A spontaneous genetically induced epiallele at a retrotransposon shapes host genome function
- PMID: 33755012
- PMCID: PMC8084528
- DOI: 10.7554/eLife.65233
A spontaneous genetically induced epiallele at a retrotransposon shapes host genome function
Abstract
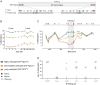
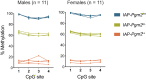
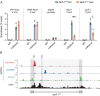
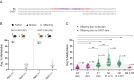
Intracisternal A-particles (IAPs) are endogenous retroviruses (ERVs) responsible for most insertional mutations in the mouse. Full-length IAPs harbour genes flanked by long terminal repeats (LTRs). Here, we identify a solo LTR IAP variant (Iap5-1solo) recently formed in the inbred C57BL/6J mouse strain. In contrast to the C57BL/6J full-length IAP at this locus (Iap5-1full), Iap5-1solo lacks DNA methylation and H3K9 trimethylation. The distinct DNA methylation levels between the two alleles are established during preimplantation development, likely due to loss of KRAB zinc finger protein binding at the Iap5-1solo variant. Iap5-1solo methylation increases and becomes more variable in a hybrid genetic background yet is unresponsive to maternal dietary methyl supplementation. Differential epigenetic modification of the two variants is associated with metabolic differences and tissue-specific changes in adjacent gene expression. Our characterisation of Iap5-1 as a genetically induced epiallele with functional consequences establishes a new model to study transposable element repression and host-element co-evolution.
Keywords: DNA methylation; IAP; chromosomes; endogenous retrovirus; epiallele; gene expression; genetics; genomics; mouse; transposable element.
Plain language summary
Our genome provides a complete set of genetic instructions for life. It begins by directing the growth and development of the embryo, and subsequently supports all the cells of the adult body in their daily routines. Yet approximately 10% of the DNA in mammalian genomes is made up of sequences originating from past retroviral infections, leaving a calling card in our genetic code. While these segments of retroviral DNA can no longer produce new infectious viruses, some of them retain the ability to copy themselves and jump into new parts of the genome. This can be problematic if they jump into and disrupt an important piece of genetic code. To protect against this, our bodies have evolved the ability to chemically strap down retroviral sequences by adding methyl groups to them and by modifying the proteins they are wrapped around. However, some of these endogenous retroviruses can dodge such so-called epigenetic modifications and disrupt genome function as a result. Studying a population of widely used inbred laboratory mice, Bertozzi et al. have identified a retroviral element that evades these epigenetic restraints. They discovered that some mice carry a full-length retroviral sequence while others have a shortened version of the same element. The shorter sequence lacked the repressive epigenetic marks found on the longer version, and this affected the expression of nearby genes. Moreover, the repressive marks could be partially restored by breeding the short-version mice with a distantly related mouse strain. Bertozzi et al. highlight an important issue for research using mouse models. Inbred laboratory mouse strains are assumed to have a fixed genetic code which allows scientists to conclude that any observed differences in their experiments are not a product of background genetic variation. However, this study emphasizes that this assumption is not guaranteed, and that hidden genetic diversity may be present in ostensibly genetically identical mice, with important implications for experimental outcomes. In addition, Bertozzi et al. provide a new mouse model for researchers to study the evolution and regulation of retroviral sequences and the impact of these processes on cell function.
© 2021, Bertozzi et al.
Conflict of interest statement
TB, NT, GH, AK, AF No competing interests declared
Figures







References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- 210757/Z/18/Z/WT_/Wellcome Trust/United Kingdom
- BB/G020930/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/R009996/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MR/R009791/1/MRC_/Medical Research Council/United Kingdom
- MR/W003783/1/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

