LIN28B induces a differentiation program through CDX2 in colon cancer
- PMID: 33755595
- PMCID: PMC8262288
- DOI: 10.1172/jci.insight.140382
LIN28B induces a differentiation program through CDX2 in colon cancer
Abstract
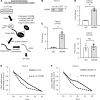
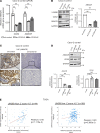
Most colorectal cancers (CRCs) are moderately differentiated or well differentiated, a status that is preserved even in metastatic tumors. However, the molecular mechanisms underlying CRC differentiation remain to be elucidated. Herein, we unravel a potentially novel posttranscriptional regulatory mechanism via a LIN28B/CDX2 signaling axis that plays a critical role in mediating CRC differentiation. Owing to a large number of mRNA targets, the mRNA-binding protein LIN28B has diverse functions in development, metabolism, tissue regeneration, and tumorigenesis. Our RNA-binding protein IP (RIP) assay revealed that LIN28B directly binds CDX2 mRNA, which is a pivotal homeobox transcription factor in normal intestinal epithelial cell identity and differentiation. Furthermore, LIN28B overexpression resulted in enhanced CDX2 expression to promote differentiation in subcutaneous xenograft tumors generated from CRC cells and metastatic tumor colonization through mesenchymal-epithelial transition in CRC liver metastasis mouse models. A ChIP sequence for CDX2 identified α-methylacyl-CoA racemase (AMACR) as a potentially novel transcriptional target of CDX2 in the context of LIN28B overexpression. We also found that AMACR enhanced intestinal alkaline phosphatase activity, which is known as a key component of intestinal differentiation, through the upregulation of butyric acid. Overall, we demonstrated that LIN28B promotes CRC differentiation through the CDX2/AMACR axis.
Keywords: Cell Biology; Colorectal cancer; Gastroenterology; Molecular biology.
Conflict of interest statement
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

