Whole-genome sequencing of SARS-CoV-2 reveals the detection of G614 variant in Pakistan
- PMID: 33755704
- PMCID: PMC7987156
- DOI: 10.1371/journal.pone.0248371
Whole-genome sequencing of SARS-CoV-2 reveals the detection of G614 variant in Pakistan
Abstract
Since its emergence in China, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has spread worldwide including Pakistan. During the pandemic, whole genome sequencing has played an important role in understanding the evolution and genomic diversity of SARS-CoV-2. Although an unprecedented number of SARS-CoV-2 full genomes have been submitted in GISAID and NCBI, data from Pakistan is scarce. We report the sequencing, genomic characterization, and phylogenetic analysis of five SARS-CoV-2 strains isolated from patients in Pakistan. The oropharyngeal swabs of patients that were confirmed positive for SARS-CoV-2 through real-time RT-PCR at National Institute of Health, Pakistan, were selected for whole-genome sequencing. Sequencing was performed using NEBNext Ultra II Directional RNA Library Prep kit for Illumina (NEW ENGLAND BioLabs Inc., MA, US) and Illumina iSeq 100 instrument (Illumina, San Diego, US). Based on whole-genome analysis, three Pakistani SARS-CoV-2 strains clustered into the 20A (GH) clade along with the strains from Oman, Slovakia, United States, and Pakistani strain EPI_ISL_513925. The two 19B (S)-clade strains were closely related to viruses from India and Oman. Overall, twenty-nine amino acid mutations were detected in the current study genome sequences, including fifteen missense and four novel mutations. Notably, we have found a D614G (aspartic acid to glycine) mutation in spike protein of the sequences from the GH clade. The G614 variant carrying the characteristic D614G mutation has been shown to be more infectious that lead to its rapid spread worldwide. This report highlights the detection of GH and S clade strains and G614 variant from Pakistan warranting large-scale whole-genome sequencing of strains prevalent in different regions to understand virus evolution and to explore their genetic diversity.
Conflict of interest statement
The authors have declared that no competing interests exist.
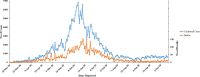
Figures


Similar articles
-
A Founder Effect Led Early SARS-CoV-2 Transmission in Spain.J Virol. 2021 Jan 13;95(3):e01583-20. doi: 10.1128/JVI.01583-20. Print 2021 Jan 13. J Virol. 2021. PMID: 33127745 Free PMC article.
-
Molecular epidemiology of SARS-CoV-2 isolated from COVID-19 family clusters.BMC Med Genomics. 2021 Jun 1;14(1):144. doi: 10.1186/s12920-021-00990-3. BMC Med Genomics. 2021. PMID: 34074255 Free PMC article.
-
Higher entropy observed in SARS-CoV-2 genomes from the first COVID-19 wave in Pakistan.PLoS One. 2021 Aug 31;16(8):e0256451. doi: 10.1371/journal.pone.0256451. eCollection 2021. PLoS One. 2021. PMID: 34464419 Free PMC article.
-
Characterization of SARS-CoV-2 different variants and related morbidity and mortality: a systematic review.Eur J Med Res. 2021 Jun 8;26(1):51. doi: 10.1186/s40001-021-00524-8. Eur J Med Res. 2021. PMID: 34103090 Free PMC article.
-
The virological impacts of SARS-CoV-2 D614G mutation.J Mol Cell Biol. 2021 Dec 30;13(10):712-720. doi: 10.1093/jmcb/mjab045. J Mol Cell Biol. 2021. PMID: 34289053 Free PMC article. Review.
Cited by
-
An Observational Study of Fungal Infections in COVID-19: Highlighting the Role of Mucormycosis in Tertiary Healthcare Settings.Cureus. 2024 Mar 30;16(3):e57295. doi: 10.7759/cureus.57295. eCollection 2024 Mar. Cureus. 2024. PMID: 38690487 Free PMC article.
-
The D614G Virus Mutation Enhances Anosmia in COVID-19 Patients: Evidence from a Systematic Review and Meta-analysis of Studies from South Asia.ACS Chem Neurosci. 2021 Oct 6;12(19):3535-3549. doi: 10.1021/acschemneuro.1c00542. Epub 2021 Sep 17. ACS Chem Neurosci. 2021. PMID: 34533304 Free PMC article.
-
Evidence of rapid rise in population immunity from SARS-CoV-2 subclinical infections through pre-vaccination serial serosurveys in Pakistan.J Glob Health. 2025 Feb 21;15:04078. doi: 10.7189/jogh.15.04078. J Glob Health. 2025. PMID: 39977666 Free PMC article.
-
Evaluation of Variant-Specific Peptides for Detection of SARS-CoV-2 Variants of Concern.J Proteome Res. 2022 Oct 7;21(10):2443-2452. doi: 10.1021/acs.jproteome.2c00325. Epub 2022 Sep 15. J Proteome Res. 2022. PMID: 36108102 Free PMC article.
-
Dual-Domain Reporter Approach for Multiplex Identification of Major SARS-CoV-2 Variants of Concern in a Microarray-Based Assay.Biosensors (Basel). 2023 Feb 13;13(2):269. doi: 10.3390/bios13020269. Biosensors (Basel). 2023. PMID: 36832035 Free PMC article.
References
-
- World Health Organization. Weekly Operational Update on COVID-19, October 16, 2020. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situatio....
-
- Pakistan confirms first two cases of coronavirus. https://www.france24.com/en/20200226-pakistan-confirms-first-two-cases-o....
-
- Tai W, He L, Zhang X, Pu J, Voronin D, et al.. (2020) Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cellular & molecular immunology 17: 613–620. 10.1038/s41423-020-0400-4 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

