mTORC1 stimulates cell growth through SAM synthesis and m6A mRNA-dependent control of protein synthesis
- PMID: 33756106
- PMCID: PMC8141029
- DOI: 10.1016/j.molcel.2021.03.009
mTORC1 stimulates cell growth through SAM synthesis and m6A mRNA-dependent control of protein synthesis
Abstract
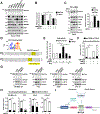
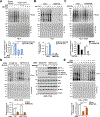
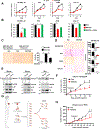
The mechanistic target of rapamycin complex 1 (mTORC1) regulates metabolism and cell growth in response to nutrient, growth, and oncogenic signals. We found that mTORC1 stimulates the synthesis of the major methyl donor, S-adenosylmethionine (SAM), through the control of methionine adenosyltransferase 2 alpha (MAT2A) expression. The transcription factor c-MYC, downstream of mTORC1, directly binds to intron 1 of MAT2A and promotes its expression. Furthermore, mTORC1 increases the protein abundance of Wilms' tumor 1-associating protein (WTAP), the positive regulatory subunit of the human N6-methyladenosine (m6A) RNA methyltransferase complex. Through the control of MAT2A and WTAP levels, mTORC1 signaling stimulates m6A RNA modification to promote protein synthesis and cell growth. A decline in intracellular SAM levels upon MAT2A inhibition decreases m6A RNA modification, protein synthesis rate, and tumor growth. Thus, mTORC1 adjusts m6A RNA modification through the control of SAM and WTAP levels to prime the translation machinery for anabolic cell growth.
Keywords: Cell growth; MAT2A; Methionine cycle; N(6)-methyladenosine; Protein Synthesis; RNA metabolism; S-adenosylmethionine; WTAP; mTOR; mTORC1.
Copyright © 2021 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




Comment in
-
More writing: mTORC1 promotes m6A mRNA methylation.Mol Cell. 2021 May 20;81(10):2057-2058. doi: 10.1016/j.molcel.2021.04.020. Mol Cell. 2021. PMID: 34019785
References
-
- Ables GP, and Johnson JE (2017). Pleiotropic responses to methionine restriction. Exp Gerontol 94, 83–88. - PubMed
-
- Adams JR, Xu K, Liu JC, Agamez NM, Loch AJ, Wong RG, Wang W, Wright KL, Lane TF, Zacksenhaus E, et al. (2011). Cooperation between Pik3ca and p53 mutations in mouse mammary tumor formation. Cancer research 71, 2706–2717. - PubMed
-
- Albers E (2009). Metabolic characteristics and importance of the universal methionine salvage pathway recycling methionine from 5’-methylthioadenosine. IUBMB Life 61, 1132–1142. - PubMed
-
- Atochina-Vasserman EN, Abramova E, James ML, Rue R, Liu AY, Ersumo NT, Guo CJ, Gow AJ, and Krymskaya VP (2015). Pharmacological targeting of VEGFR signaling with axitinib inhibits Tsc2-null lesion growth in the mouse model of lymphangioleiomyomatosis. Am J Physiol Lung Cell Mol Physiol 309, L1447–1454. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

