The antigenic anatomy of SARS-CoV-2 receptor binding domain
- PMID: 33756110
- PMCID: PMC7891125
- DOI: 10.1016/j.cell.2021.02.032
The antigenic anatomy of SARS-CoV-2 receptor binding domain
Abstract
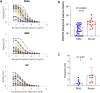
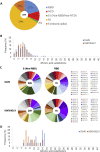
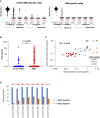
Antibodies are crucial to immune protection against SARS-CoV-2, with some in emergency use as therapeutics. Here, we identify 377 human monoclonal antibodies (mAbs) recognizing the virus spike and focus mainly on 80 that bind the receptor binding domain (RBD). We devise a competition data-driven method to map RBD binding sites. We find that although antibody binding sites are widely dispersed, neutralizing antibody binding is focused, with nearly all highly inhibitory mAbs (IC50 < 0.1 μg/mL) blocking receptor interaction, except for one that binds a unique epitope in the N-terminal domain. Many of these neutralizing mAbs use public V-genes and are close to germline. We dissect the structural basis of recognition for this large panel of antibodies through X-ray crystallography and cryoelectron microscopy of 19 Fab-antigen structures. We find novel binding modes for some potently inhibitory antibodies and demonstrate that strongly neutralizing mAbs protect, prophylactically or therapeutically, in animal models.
Keywords: coronavirus, SARS-CoV-2, anti-RBD antibody, receptor binding domain, antibody, immune responses, virus structure.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests M.S.D. is a consultant for Inbios, Vir Biotechnology, NGM Biopharmaceuticals, and Carnival Corporation and is on the Scientific Advisory Boards of Moderna and Immunome. The M.S.D. laboratory has received unrelated funding support in sponsored research agreements from Moderna, Vir Biotechnology, and Emergent BioSolutions. G.R.S. sits on the GSK Vaccines Scientific Advisory Board. A.J.P. is Chair of UK Department Health and Social Care’s (DHSC) Joint Committee on Vaccination & Immunisation (JCVI), but does not chair or participate in the JCVI COVID19 committee, and is a member of the WHO’s SAGE. The views expressed in this article do not necessarily represent the views of DHSC, JCVI, NIHR, or WHO. The University of Oxford has entered into a partnership with AstraZeneca on coronavirus vaccine development. All other authors declare no competing financial interests. The University of Oxford has protected intellectual property disclosed in this publication.
Figures










References
-
- Aricescu A.R., Lu W., Jones E.Y. A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr Sect D Biol Crystallogr. 2006;62:1243–1250. - PubMed
-
- Barnes C.O., West A.P., Jr., Huey-Tubman K.E., Hoffmann M.A.G., Sharaf N.G., Hoffman P.R., Koranda N., Gristick H.B., Gaebler C., Muecksch F. Structures of Human Antibodies Bound to SARS-CoV-2 Spike Reveal Common Epitopes and Recurrent Features of Antibodies. Cell. 2020;182:828–842.e16. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 203224/Z/16/Z/WT_/Wellcome Trust/United Kingdom
- R01 AI157155/AI/NIAID NIH HHS/United States
- MR/V001329/1/MRC_/Medical Research Council/United Kingdom
- MC_UU_00008/11/MRC_/Medical Research Council/United Kingdom
- MC_PC_17137/MRC_/Medical Research Council/United Kingdom
- MC_PC_20016/MRC_/Medical Research Council/United Kingdom
- MR/L018942/1/MRC_/Medical Research Council/United Kingdom
- MC_PC_19060/MRC_/Medical Research Council/United Kingdom
- G0600520/MRC_/Medical Research Council/United Kingdom
- MR/N00065X/1/MRC_/Medical Research Council/United Kingdom
- T32 AI007172/AI/NIAID NIH HHS/United States
- 109965/Z/15/Z/WT_/Wellcome Trust/United Kingdom
- G1001046/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

