A region-based method for causal mediation analysis of DNA methylation data
- PMID: 33757385
- PMCID: PMC8920164
- DOI: 10.1080/15592294.2021.1900026
A region-based method for causal mediation analysis of DNA methylation data
Abstract
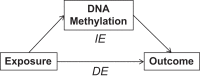
Exposure to environmental factors can affect DNA methylation at a 5'-cytosine-phosphate-guanine-3' (CpG) site or a genomic region, which can then affect an outcome. In other words, environmental effects on an outcome could be mediated by DNA methylation. To date, single CpG-site-based mediation analysis has been employed extensively. More recently, however, there has been considerable interest in studying differentially methylated regions (DMRs), both because DMRs are more likely to have functional effects than single CpG sites and because testing DMRs reduces multiple testing. In this report, we propose a novel causal mediation approach under the counterfactual framework to test the significance of total (TE), direct (DE), and indirect effects (IE) of predictors on response variable with a methylated region (MR) as the mediator (denoted as MR-Mediation). Functional linear transformation is used to reduce the possible high dimension of the CpG sites in a predefined MR and to account for their location information. In our simulation studies, MR-Mediation retained the desired Type I error rates for TE, DE, and IE tests. Furthermore, MR-Mediation had better power performance than testing mean methylation level as the mediator in most considered scenarios, especially for IE (i.e., mediated effect) test, which could be more interesting than the other two effect tests. We further illustrate our proposed method by analysing the methylation mediated effect of exposure to gun violence on total immunoglobulin E or atopic asthma among participants in the Epigenetic Variation and Childhood Asthma in Puerto Ricans study.
Keywords: Counterfactual framework; Mediation; Methylated region.
Figures


References
-
- Portela A, Esteller M.. Epigenetic modifications and human disease. Nat Biotechnol. 2010;28:1057–1068. - PubMed
-
- Melotte V, Lentjes MH, Van Den Bosch SM, et al. N-Myc downstream-regulated gene 4 (NDRG4): a candidate tumor suppressor gene and potential biomarker for colorectal cancer. J Natl Cancer Inst. 2009;101:916–927. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
