A genome-scale CRISPR interference guide library enables comprehensive phenotypic profiling in yeast
- PMID: 33757429
- PMCID: PMC7986282
- DOI: 10.1186/s12864-021-07518-0
A genome-scale CRISPR interference guide library enables comprehensive phenotypic profiling in yeast
Abstract
Background: CRISPR/Cas9-mediated transcriptional interference (CRISPRi) enables programmable gene knock-down, yielding loss-of-function phenotypes for nearly any gene. Effective, inducible CRISPRi has been demonstrated in budding yeast, and genome-scale guide libraries enable systematic, genome-wide genetic analysis.
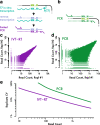
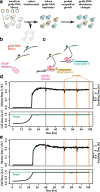
Results: We present a comprehensive yeast CRISPRi library, based on empirical design rules, containing 10 distinct guides for most genes. Competitive growth after pooled transformation revealed strong fitness defects for most essential genes, verifying that the library provides comprehensive genome coverage. We used the relative growth defects caused by different guides targeting essential genes to further refine yeast CRISPRi design rules. In order to obtain more accurate and robust guide abundance measurements in pooled screens, we link guides with random nucleotide barcodes and carry out linear amplification by in vitro transcription.
Conclusions: Taken together, we demonstrate a broadly useful platform for comprehensive, high-precision CRISPRi screening in yeast.
Keywords: Budding yeast; CRISPR interference; Pooled screening.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

