Analysis of the effects of related fingerprints on molecular similarity using an eigenvalue entropy approach
- PMID: 33757582
- PMCID: PMC7989080
- DOI: 10.1186/s13321-021-00506-2
Analysis of the effects of related fingerprints on molecular similarity using an eigenvalue entropy approach
Abstract
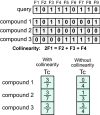
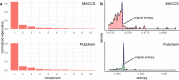
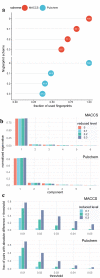
Two-dimensional (2D) chemical fingerprints are widely used as binary features for the quantification of structural similarity of chemical compounds, which is an important step in similarity-based virtual screening (VS). Here, using an eigenvalue-based entropy approach, we identified 2D fingerprints with little to no contribution to shaping the eigenvalue distribution of the feature matrix as related ones and examined the degree to which these related 2D fingerprints influenced molecular similarity scores calculated with the Tanimoto coefficient. Our analysis identified many related fingerprints in publicly available fingerprint schemes and showed that their presence in the feature set could have substantial effects on the similarity scores and bias the outcome of molecular similarity analysis. Our results have implication in the optimal selection of 2D fingerprints for compound similarity analysis and the identification of potential hits for compounds with target biological activity in VS.
Keywords: 2D fingerprint; Chemoinformatics; Similarity-based virtual screening; Structure-activity relationship; Unsupervised feature selection.
Conflict of interest statement
We declare that we have no competing interests.
Figures





Similar articles
-
How do 2D fingerprints detect structurally diverse active compounds? Revealing compound subset-specific fingerprint features through systematic selection.J Chem Inf Model. 2011 Sep 26;51(9):2254-65. doi: 10.1021/ci200275m. Epub 2011 Aug 8. J Chem Inf Model. 2011. PMID: 21793563
-
Modeling Tanimoto Similarity Value Distributions and Predicting Search Results.Mol Inform. 2017 Jul;36(7). doi: 10.1002/minf.201600131. Epub 2016 Dec 29. Mol Inform. 2017. PMID: 28032955
-
Database fingerprint (DFP): an approach to represent molecular databases.J Cheminform. 2017 Feb 6;9:9. doi: 10.1186/s13321-017-0195-1. eCollection 2017. J Cheminform. 2017. PMID: 28224019 Free PMC article.
-
Similarity-based virtual screening using 2D fingerprints.Drug Discov Today. 2006 Dec;11(23-24):1046-53. doi: 10.1016/j.drudis.2006.10.005. Epub 2006 Oct 20. Drug Discov Today. 2006. PMID: 17129822 Review.
-
Mini-fingerprints for virtual screening: design principles and generation of novel prototypes based on information theory.SAR QSAR Environ Res. 2003 Feb;14(1):27-40. doi: 10.1080/1062936021000058764. SAR QSAR Environ Res. 2003. PMID: 12688414 Review.
Cited by
-
LCK-SafeScreen-Model: An Advanced Ensemble Machine Learning Approach for Estimating the Binding Affinity between Compounds and LCK Target.Molecules. 2023 Nov 1;28(21):7382. doi: 10.3390/molecules28217382. Molecules. 2023. PMID: 37959801 Free PMC article.
-
Exploring Natural Compounds as Potential CDK4 Inhibitors for Therapeutic Intervention in Neurodegenerative Diseases through Computational Analysis.Mol Biotechnol. 2025 Aug;67(8):3310-3329. doi: 10.1007/s12033-024-01258-8. Epub 2024 Aug 29. Mol Biotechnol. 2025. PMID: 39207668
-
DeepSAT: Learning Molecular Structures from Nuclear Magnetic Resonance Data.J Cheminform. 2023 Aug 7;15(1):71. doi: 10.1186/s13321-023-00738-4. J Cheminform. 2023. PMID: 37550756 Free PMC article.
-
Chemistry-informed recommender system to predict optimal molecular receptors in SERS nanosensors.Nat Commun. 2025 Aug 2;16(1):7095. doi: 10.1038/s41467-025-62519-x. Nat Commun. 2025. PMID: 40753173 Free PMC article.
-
The Goldilocks paradigm: comparing classical machine learning, large language models, and few-shot learning for drug discovery applications.Commun Chem. 2024 Jun 12;7(1):134. doi: 10.1038/s42004-024-01220-4. Commun Chem. 2024. PMID: 38866916 Free PMC article.
References
-
- Smith A. Screening for drug discovery: the leading question. Nature. 2002;418:453–459. - PubMed
Grants and funding
- BAS/1/1624-01/King Abdullah University of Science and Technology
- URF/1/3412-01/King Abdullah University of Science and Technology
- URF/1/3450-01/King Abdullah University of Science and Technology
- FCC/1/1976-18/King Abdullah University of Science and Technology
- FCC/1/1976-23/King Abdullah University of Science and Technology
LinkOut - more resources
Full Text Sources
Other Literature Sources

