Genetic relatedness of multidrug resistant Escherichia coli isolated from humans, chickens and poultry environments
- PMID: 33757589
- PMCID: PMC7988975
- DOI: 10.1186/s13756-021-00930-x
Genetic relatedness of multidrug resistant Escherichia coli isolated from humans, chickens and poultry environments
Abstract
Background: Inappropriate use of antimicrobial agents in animal production has led to the development of antimicrobial resistance (AMR) in foodborne pathogens. Transmission of AMR foodborne pathogens from reservoirs, particularly chickens to the human population does occur. Recently, we reported that occupational exposure was a risk factor for multidrug-resistant (MDR) Escherichia coli (E. coli) among poultry-workers. Here we determined the prevalence and genetic relatedness among MDR E. coli isolated from poultry-workers, chickens, and poultry environments in Abuja, Nigeria. This study was conducted to address the gaps identified by the Nigerian AMR situation analysis.
Methods: We conducted a cross-sectional study among poultry-workers, chickens, and poultry farm/live bird market (LBM) environments. The isolates were tested phenotypically for their antimicrobial susceptibility profiles, genotypically characterized using whole-genome sequencing (WGS) and in silico multilocus sequence types (MLST). We conducted a phylogenetic single nucleotide polymorphism (SNPs) analysis to determine relatedness and clonality among the isolates.
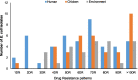
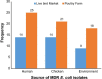
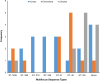
Results: A total of 115 (26.8%) out of 429 samples were positive for E. coli. Of these, 110 isolates were viable for phenotypic and genotypic characterization. The selection comprised 47 (42.7%) isolates from poultry-workers, 36 (32.7%) from chickens, and 27 (24.5%) from poultry-farm or LBM environments. Overall, 101 (91.8%) of the isolates were MDR conferring resistance to at least three drug classes. High frequency of resistance was observed for tetracycline (n = 102; 92.7%), trimethoprim/sulfamethoxazole (n = 93; 84.5%), streptomycin (n = 87; 79.1%) and ampicillin (n = 88; 80%). Two plasmid-mediated colistin genes-mcr-1.1 harboured on IncX4 plasmids were detected in environmental isolates. The most prevalent sequence types (ST) were ST-155 (n = 8), ST-48 (n = 8) and ST-10 (n = 6). Two isolates of human and environmental sources with a SNPs difference of 6161 originating from the same farm shared a novel ST. The isolates had similar AMR genes and plasmid replicons.
Conclusion: MDR E.coli isolates were prevalent amongst poultry-workers, poultry, and the poultry farm/LBM environment. The emergence of MDR E. coli with novel ST in two isolates may be plasmid-mediated. Competent authorities should enforce AMR regulations to ensure prudent use of antimicrobials to limit the risk of transmission along the food chain.
Keywords: Antimicrobial resistance; Chicken; Escherichia coli; Genetic relatedness; Multidrug resistance; Nigeria; One health; Prevalence.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Parmley J, Leung Z, Léger D, Finley R, Irwin R, Pintar K, et al. One Health and Food Safety—The Canadian Experience: A Holistic approach toward enteric bacteria pathogens and antimicrobial resistance surveillance. Natl Acad Press. 2012. Available online at https://www.ncbi.nlm.nih.gov/books/NBK114511/.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

