A Novel Metabolism-Related Signature as a Candidate Prognostic Biomarker for Hepatocellular Carcinoma
- PMID: 33758763
- PMCID: PMC7981163
- DOI: 10.2147/JHC.S294108
A Novel Metabolism-Related Signature as a Candidate Prognostic Biomarker for Hepatocellular Carcinoma
Abstract
Purpose: Given that metabolic reprogramming has been recognized as an essential hallmark of cancer cells, this study sought to investigate the potential prognostic values of metabolism-related genes (MRGs) for the diagnosis and treatment of hepatocellular carcinoma (HCC).
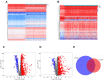
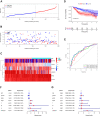
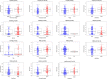
Methods: In total, 2752 metabolism-related gene sequencing data of HCC samples with clinical information were obtained from the International Cancer Genome Consortium (ICGC) and The Cancer Genome Atlas (TCGA). One hundred and seventy-eight the differentially expressed MRGs were identified from the ICGC cohort and TCGA cohort. Then, univariate Cox regression analysis was performed to identify these genes that were related to overall survival (OS). A novel metabolism-related prognostic signature was developed using the least absolute shrinkage and selection operator (Lasso) and multivariate Cox regression analyses in the ICGC dataset. The Broad Institute's Connectivity Map (CMap) was used in predicting which compounds on the basis of the prognostic MRGs. Furthermore, the signature was validated in the TCGA dataset. Finally, the expression levels of hub genes were validated in HCC cell lines by Western blotting (WB) and quantitative real-time PCR (qRT-PCR).
Results: We found that 17 MRGs were most significantly associated with OS in HCC. Then, the Lasso and multivariate Cox regression analyses were applied to construct the novel metabolism-relevant prognostic signature, which consisted of six MRGs. The prognostic value of this prognostic model was further successfully validated in the TCGA dataset. Further analysis indicated that this particular signature could be an independent prognostic indicator after adjusting to other clinical factors. Six MRGs (FLVCR1, MOGAT2, SLC5A11, RRM2, COX7B2, and SCN4A) showed high prognostic performance in predicting HCC outcomes. Candidate drugs that aimed at hub ERGs were identified. Finally, hub genes were chosen for validation and the protein, mRNA expression of FLVCR1, SLC5A11, and RRM2 were significantly increased in human HCC cell lines compared to normal human hepatic cell lines, which were in agreement with the results of differential expression analysis.
Conclusion: Our data provided evidence that the metabolism-related signature could serve as a reliable prognostic and predictive tool for OS in patients with HCC.
Keywords: biomarker; hepatocellular carcinoma; metabolism-related genes; prognostic signature.
© 2021 Wang et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest for this work.
Figures






References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

