This is a preprint.
A humanized mouse model of chronic COVID-19 to evaluate disease mechanisms and treatment options
- PMID: 33758831
- PMCID: PMC7987100
- DOI: 10.21203/rs.3.rs-279341/v1
A humanized mouse model of chronic COVID-19 to evaluate disease mechanisms and treatment options
Update in
-
A humanized mouse model of chronic COVID-19.Nat Biotechnol. 2022 Jun;40(6):906-920. doi: 10.1038/s41587-021-01155-4. Epub 2021 Dec 17. Nat Biotechnol. 2022. PMID: 34921308 Free PMC article.
Abstract
Coronavirus-associated acute respiratory disease, called coronavirus disease 2019 (COVID-19) is an infectious disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). More than 90 million people have been infected with SARS-CoV-2 and more than 2 million people have died of complications due to COVID-19 worldwide. COVID-19, in its severe form, presents with an uncontrolled, hyperactive immune response and severe immunological injury or organ damage that accounts for morbidity and mortality. Even in the absence of complications, COVID-19 can last for several months with lingering effects of an overactive immune system. Dysregulated myeloid and lymphocyte compartments have been implicated in lung immunopathology. Currently, there are limited clinically-tested treatments of COVID-19 with disparities in the apparent efficacy in patients. Accurate model systems are essential to rapidly evaluate promising discoveries but most currently available in mice, ferrets and hamsters do not recapitulate sustained immunopathology described in COVID19 patients. Here, we present a comprehensively humanized mouse COVID-19 model that faithfully recapitulates the innate and adaptive human immune responses during infection with SARS-CoV-2 by adapting recombinant adeno-associated virus (AAV)-driven gene therapy to deliver human ACE2 to the lungs 1 of MISTRG6 mice. Our unique model allows for the first time the study of chronic disease due to infection with SARS-CoV-2 in the context of patient-derived antibodies to characterize in real time the potential culprits of the observed human driving immunopathology; most importantly this model provides a live view into the aberrant macrophage response that is thought to be the effector of disease morbidity and ARDS in patients. Application of therapeutics such as patient-derived antibodies and steroids to our model allowed separation of the two aspects of the immune response, infectious viral clearance and immunopathology. Inflammatory cells seeded early in infection drove immune-patholgy later, but this very same early anti-viral response was also crucial to contain infection.
Conflict of interest statement
Competing financial interests
The authors declare no competing financial interests.
Figures
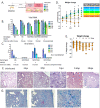
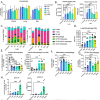


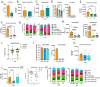

References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

