This is a preprint.
The N501Y spike substitution enhances SARS-CoV-2 transmission
- PMID: 33758836
- PMCID: PMC7986995
- DOI: 10.1101/2021.03.08.434499
The N501Y spike substitution enhances SARS-CoV-2 transmission
Update in
-
The N501Y spike substitution enhances SARS-CoV-2 infection and transmission.Nature. 2022 Feb;602(7896):294-299. doi: 10.1038/s41586-021-04245-0. Epub 2021 Nov 24. Nature. 2022. PMID: 34818667 Free PMC article.
Abstract
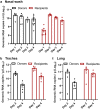
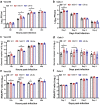
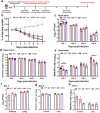
Beginning in the summer of 2020, a variant of SARS-CoV-2, the cause of the COVID-19 pandemic, emerged in the United Kingdom (UK). This B.1.1.7 variant increased rapidly in prevalence among sequenced strains, attributed to an increase in infection and/or transmission efficiency. The UK variant has 19 nonsynonymous mutations across its viral genome including 8 substitutions or deletions in the spike protein, which interacts with cellular receptors to mediate infection and tropism. Here, using a reverse genetics approach, we show that, of the 8 individual spike protein substitutions, only N501Y exhibited consistent fitness gains for replication in the upper airway in the hamster model as well as primary human airway epithelial cells. The N501Y substitution recapitulated the phenotype of enhanced viral transmission seen with the combined 8 UK spike mutations, suggesting it is a major determinant responsible for increased transmission of this variant. Mechanistically, the N501Y substitution improved the affinity of the viral spike protein for cellular receptors. As suggested by its convergent evolution in Brazil and South Africa, our results indicate that N501Y substitution is a major adaptive spike mutation of major concern.
Conflict of interest statement
Competing financial interests
X.X., V.D.M., and P.-Y.S. have filed a patent on the reverse genetic system and reporter SARS-CoV-2. Other authors declare no competing interests.
Figures






References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
