This is a preprint.
Elicitation of broadly protective sarbecovirus immunity by receptor-binding domain nanoparticle vaccines
- PMID: 33758839
- PMCID: PMC7986998
- DOI: 10.1101/2021.03.15.435528
Elicitation of broadly protective sarbecovirus immunity by receptor-binding domain nanoparticle vaccines
Update in
-
Elicitation of broadly protective sarbecovirus immunity by receptor-binding domain nanoparticle vaccines.Cell. 2021 Oct 14;184(21):5432-5447.e16. doi: 10.1016/j.cell.2021.09.015. Epub 2021 Sep 15. Cell. 2021. PMID: 34619077 Free PMC article.
Abstract
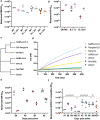
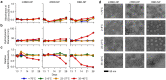
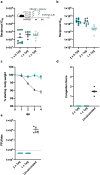
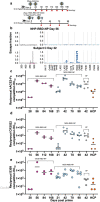
Understanding the ability of SARS-CoV-2 vaccine-elicited antibodies to neutralize and protect against emerging variants of concern and other sarbecoviruses is key for guiding vaccine development decisions and public health policies. We show that a clinical stage multivalent SARS-CoV-2 receptor-binding domain nanoparticle vaccine (SARS-CoV-2 RBD-NP) protects mice from SARS-CoV-2-induced disease after a single shot, indicating that the vaccine could allow dose-sparing. SARS-CoV-2 RBD-NP elicits high antibody titers in two non-human primate (NHP) models against multiple distinct RBD antigenic sites known to be recognized by neutralizing antibodies. We benchmarked NHP serum neutralizing activity elicited by RBD-NP against a lead prefusion-stabilized SARS-CoV-2 spike immunogen using a panel of single-residue spike mutants detected in clinical isolates as well as the B.1.1.7 and B.1.351 variants of concern. Polyclonal antibodies elicited by both vaccines are resilient to most RBD mutations tested, but the E484K substitution has similar negative consequences for neutralization, and exhibit modest but comparable neutralization breadth against distantly related sarbecoviruses. We demonstrate that mosaic and cocktail sarbecovirus RBD-NPs elicit broad sarbecovirus neutralizing activity, including against the SARS-CoV-2 B.1.351 variant, and protect mice against severe SARS-CoV challenge even in the absence of the SARS-CoV RBD in the vaccine. This study provides proof of principle that sarbecovirus RBD-NPs induce heterotypic protection and enables advancement of broadly protective sarbecovirus vaccines to the clinic.
Conflict of interest statement
Declaration of interest A.C.W, N.P.K. and D.V., are named as inventors on patent applications filed by the University of Washington based on the studies presented in this paper. N.P.K. is a co-founder, shareholder, paid consultant, and chair of the scientific advisory board of Icosavax, Inc. and has received an unrelated sponsored research agreement from Pfizer. D.V. is a consultant for and has received an unrelated sponsored research agreement from Vir Biotechnology Inc. R.R., D.T.O., and R.V.D.M are employees of GlaxoSmithKline. C.-L.H. and J.S.M. are inventors on U.S. patent application no. 63/032,502 “Engineered Coronavirus Spike (S) Protein and Methods of Use Thereof. The other authors declare no competing interests.
Figures



References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
