This is a preprint.
Interferon Resistance of Emerging SARS-CoV-2 Variants
- PMID: 33758840
- PMCID: PMC7986999
- DOI: 10.1101/2021.03.20.436257
Interferon Resistance of Emerging SARS-CoV-2 Variants
Update in
-
Interferon resistance of emerging SARS-CoV-2 variants.Proc Natl Acad Sci U S A. 2022 Aug 9;119(32):e2203760119. doi: 10.1073/pnas.2203760119. Epub 2022 Jul 22. Proc Natl Acad Sci U S A. 2022. PMID: 35867811 Free PMC article.
Abstract
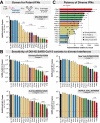
The emergence of SARS-CoV-2 variants with enhanced transmissibility, pathogenesis and resistance to vaccines presents urgent challenges for curbing the COVID-19 pandemic. While Spike mutations that enhance virus infectivity or neutralizing antibody evasion may drive the emergence of these novel variants, studies documenting a critical role for interferon responses in the early control of SARS-CoV-2 infection, combined with the presence of viral genes that limit these responses, suggest that interferons may also influence SARS-CoV-2 evolution. Here, we compared the potency of 17 different human interferons against multiple viral lineages sampled during the course of the global outbreak, including ancestral and four major variants of concern. Our data reveal increased interferon resistance in emerging SARS-CoV-2 variants, suggesting that evasion of innate immunity may be a significant, ongoing driving force for SARS-CoV-2 evolution. These findings have implications for the increased lethality of emerging variants and highlight the interferon subtypes that may be most successful in the treatment of early infections.
Figures




References
-
- Pestka S, Krause CD, Walter MR. Interferons, interferon-like cytokines, and their receptors. Immunol Rev. 2004;202:8–32. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
