This is a preprint.
Development of potency, breadth and resilience to viral escape mutations in SARS-CoV-2 neutralizing antibodies
- PMID: 33758864
- PMCID: PMC7987023
- DOI: 10.1101/2021.03.07.434227
Development of potency, breadth and resilience to viral escape mutations in SARS-CoV-2 neutralizing antibodies
Update in
-
Affinity maturation of SARS-CoV-2 neutralizing antibodies confers potency, breadth, and resilience to viral escape mutations.Immunity. 2021 Aug 10;54(8):1853-1868.e7. doi: 10.1016/j.immuni.2021.07.008. Epub 2021 Jul 30. Immunity. 2021. PMID: 34331873 Free PMC article.
Abstract
Antibodies elicited in response to infection undergo somatic mutation in germinal centers that can result in higher affinity for the cognate antigen. To determine the effects of somatic mutation on the properties of SARS-CoV-2 spike receptor-binding domain (RBD)-specific antibodies, we analyzed six independent antibody lineages. As well as increased neutralization potency, antibody evolution changed pathways for acquisition of resistance and, in some cases, restricted the range of neutralization escape options. For some antibodies, maturation apparently imposed a requirement for multiple spike mutations to enable escape. For certain antibody lineages, maturation enabled neutralization of circulating SARS-CoV-2 variants of concern and heterologous sarbecoviruses. Antibody-antigen structures revealed that these properties resulted from substitutions that allowed additional variability at the interface with the RBD. These findings suggest that increasing antibody diversity through prolonged or repeated antigen exposure may improve protection against diversifying SARS-CoV-2 populations, and perhaps against other pandemic threat coronaviruses.
Keywords: Antibodies; Neutralization; SARS-CoV-2.
Conflict of interest statement
Declaration of Interests The Rockefeller University has filed provisional patent applications in connection with this work on which M.C.N. (US patent 63/021,387) and Y.W., F.S., T.H. and P.D.B. (US patent 63/036,124) are listed as inventors.
Figures

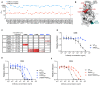
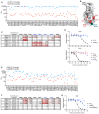




References
-
- Barnes C.O., West A.P. Jr., Huey-Tubman K.E., Hoffmann M.A.G., Sharaf N.G., Hoffman P.R., Koranda N., Gristick H.B., Gaebler C., Muecksch F., et al. (2020c). Structures of Human Antibodies Bound to SARS-CoV-2 Spike Reveal Common Epitopes and Recurrent Features of Antibodies. Cell 182, 828–842.e816. - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
