This is a preprint.
Structural basis for backtracking by the SARS-CoV-2 replication-transcription complex
- PMID: 33758867
- PMCID: PMC7987028
- DOI: 10.1101/2021.03.13.435256
Structural basis for backtracking by the SARS-CoV-2 replication-transcription complex
Update in
-
Structural Basis for Helicase-Polymerase Coupling in the SARS-CoV-2 Replication-Transcription Complex.Cell. 2020 Sep 17;182(6):1560-1573.e13. doi: 10.1016/j.cell.2020.07.033. Epub 2020 Jul 28. Cell. 2020. PMID: 32783916 Free PMC article.
-
Structural basis for backtracking by the SARS-CoV-2 replication-transcription complex.Proc Natl Acad Sci U S A. 2021 May 11;118(19):e2102516118. doi: 10.1073/pnas.2102516118. Proc Natl Acad Sci U S A. 2021. PMID: 33883267 Free PMC article.
Abstract
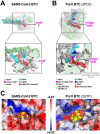
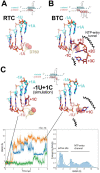
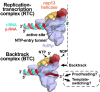
Backtracking, the reverse motion of the transcriptase enzyme on the nucleic acid template, is a universal regulatory feature of transcription in cellular organisms but its role in viruses is not established. Here we present evidence that backtracking extends into the viral realm, where backtracking by the SARS-CoV-2 RNA-dependent RNA polymerase (RdRp) may aid viral transcription and replication. Structures of SARS-CoV-2 RdRp bound to the essential nsp13 helicase and RNA suggested the helicase facilitates backtracking. We use cryo-electron microscopy, RNA-protein crosslinking, and unbiased molecular dynamics simulations to characterize SARS-CoV-2 RdRp backtracking. The results establish that the single-stranded 3'-segment of the product-RNA generated by backtracking extrudes through the RdRp NTP-entry tunnel, that a mismatched nucleotide at the product-RNA 3'-end frays and enters the NTP-entry tunnel to initiate backtracking, and that nsp13 stimulates RdRp backtracking. Backtracking may aid proofreading, a crucial process for SARS-CoV-2 resistance against antivirals.
Keywords: Biological Sciences; Biophysics and Computational Biology; RNA-dependent RNA polymerase; backtracking; coronavirus; cryo-electron microscopy; molecular dynamics.
Conflict of interest statement
Competing Interest Statement: The authors declare no conflict of interest.
Figures



References
-
- U. S. F. and D. Administration, Remdesivir-EUA-Letter-Of-Authorization.pdf. undefined (2020).
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
