This is a preprint.
Transmission, infectivity, and antibody neutralization of an emerging SARS-CoV-2 variant in California carrying a L452R spike protein mutation
- PMID: 33758899
- PMCID: PMC7987058
- DOI: 10.1101/2021.03.07.21252647
Transmission, infectivity, and antibody neutralization of an emerging SARS-CoV-2 variant in California carrying a L452R spike protein mutation
Update in
-
Transmission, infectivity, and neutralization of a spike L452R SARS-CoV-2 variant.Cell. 2021 Jun 24;184(13):3426-3437.e8. doi: 10.1016/j.cell.2021.04.025. Epub 2021 Apr 20. Cell. 2021. PMID: 33991487 Free PMC article.
Abstract
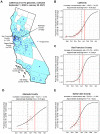
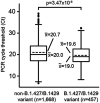
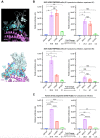
We identified a novel SARS-CoV-2 variant by viral whole-genome sequencing of 2,172 nasal/nasopharyngeal swab samples from 44 counties in California. Named B.1.427/B.1.429 to denote its 2 lineages, the variant emerged around May 2020 and increased from 0% to >50% of sequenced cases from September 1, 2020 to January 29, 2021, exhibiting an 18.6-24% increase in transmissibility relative to wild-type circulating strains. The variant carries 3 mutations in the spike protein, including an L452R substitution. Our analyses revealed 2-fold increased B.1.427/B.1.429 viral shedding in vivo and increased L452R pseudovirus infection of cell cultures and lung organoids, albeit decreased relative to pseudoviruses carrying the N501Y mutation found in the B.1.1.7, B.1.351, and P.1 variants. Antibody neutralization assays showed 4.0 to 6.7-fold and 2.0-fold decreases in neutralizing titers from convalescent patients and vaccine recipients, respectively. The increased prevalence of a more transmissible variant in California associated with decreased antibody neutralization warrants further investigation.
Figures


References
-
- Avanzato V.A., Matson M.J., Seifert S.N., Pryce R., Williamson B.N., Anzick S.L., Barbian K., Judson S.D., Fischer E.R., Martens C., et al. (2020). Case Study: Prolonged Infectious SARS-CoV-2 Shedding from an Asymptomatic Immunocompromised Individual with Cancer. Cell 183, 1901–1912 e1909. - PMC - PubMed
-
- Bedford T., Hodcroft E.B., and Neher R.A. (2021). Updated Nextstrain SARS-CoV-2 clade naming strategy (NextStrain).
-
- Bedford T., and Neher R. (2020). A Getting Started Guide to the Genomic Epidemiology of SARS-CoV-2. In Nextstrain documentation (Nextstrain.org).
-
- Bushnell B. (2021). BBMap short read aligner, and other bioinformatic tools.
-
- Buss L.F., Prete C.A. Jr., Abrahim C.M.M., Mendrone A. Jr., Salomon T., de Almeida-Neto C., Franca R.F.O., Belotti M.C., Carvalho M., Costa A.G., et al. (2021). Three-quarters attack rate of SARS-CoV-2 in the Brazilian Amazon during a largely unmitigated epidemic. Science 371, 288–292. - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
