Biased Docking for Protein-Ligand Pose Prediction
- PMID: 33759120
- PMCID: PMC10708986
- DOI: 10.1007/978-1-0716-1209-5_3
Biased Docking for Protein-Ligand Pose Prediction
Abstract
The interaction between a protein and its ligands is one of the basic and most important processes in biological chemistry. Docking methods aim to predict the molecular 3D structure of protein-ligand complexes starting from coordinates of the protein and the ligand separately. They are widely used in both industry and academia, especially in the context of drug development projects. AutoDock4 is one of the most popular docking tools and, as for any docking method, its performance is highly system dependent. Knowledge about specific protein-ligand interactions on a particular target can be used to successfully overcome this limitation. Here, we describe how to apply the AutoDock Bias protocol, a simple and elegant strategy that allows users to incorporate target-specific information through a modified scoring function that biases the ligand structure towards those poses (or conformations) that establish selected interactions. We discuss two examples using different bias sources. In the first, we show how to steer dockings towards interactions derived from crystal structures of the receptor with different ligands; in the second example, we define and apply hydrophobic biases derived from Molecular Dynamics simulations in mixed solvents. Finally, we discuss general concepts of biased docking, its performance in pose prediction, and virtual screening campaigns as well as other potential applications.
Keywords: AutoDock; AutoDock Bias; Biased docking; Cosolvent; Docking; Guided docking; Knowledge-based docking; Mixed-solvents.
Figures


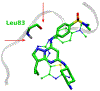






References
-
- Goodsell DS and Olson AJ (1990) Automated docking of substrates to proteins by simulated annealing. Proteins 8:195–202 - PubMed
-
- Hart TN and Read RJ (1992) A multiple-start Monte Carlo docking method. Proteins 13:206–222 - PubMed
-
- Morris GM, Goodsell DS, Halliday RS, et al. (1998) Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. 19:1639–1662
-
- Jones G, Willett P, Glen RC, et al. (1997) Development and validation of a genetic algorithm for flexible docking. J Mol Biol 267:727–748 - PubMed
-
- Friesner RA, Banks JL, Murphy RB, et al. (2004) Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem 47:1739–1749 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

