Mitochondrial metabolism supports resistance to IDH mutant inhibitors in acute myeloid leukemia
- PMID: 33760042
- PMCID: PMC7995203
- DOI: 10.1084/jem.20200924
Mitochondrial metabolism supports resistance to IDH mutant inhibitors in acute myeloid leukemia
Abstract
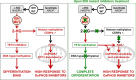
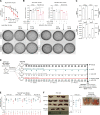
Mutations in IDH induce epigenetic and transcriptional reprogramming, differentiation bias, and susceptibility to mitochondrial inhibitors in cancer cells. Here, we first show that cell lines, PDXs, and patients with acute myeloid leukemia (AML) harboring an IDH mutation displayed an enhanced mitochondrial oxidative metabolism. Along with an increase in TCA cycle intermediates, this AML-specific metabolic behavior mechanistically occurred through the increase in electron transport chain complex I activity, mitochondrial respiration, and methylation-driven CEBPα-induced fatty acid β-oxidation of IDH1 mutant cells. While IDH1 mutant inhibitor reduced 2-HG oncometabolite and CEBPα methylation, it failed to reverse FAO and OxPHOS. These mitochondrial activities were maintained through the inhibition of Akt and enhanced activation of peroxisome proliferator-activated receptor-γ coactivator-1 PGC1α upon IDH1 mutant inhibitor. Accordingly, OxPHOS inhibitors improved anti-AML efficacy of IDH mutant inhibitors in vivo. This work provides a scientific rationale for combinatory mitochondrial-targeted therapies to treat IDH mutant AML patients, especially those unresponsive to or relapsing from IDH mutant inhibitors.
© 2021 Stuani et al.
Conflict of interest statement
Disclosures: B. Nicolay reported "other" from Agios Pharmaceuticals outside the submitted work and is an employee and shareholder of Agios Pharmaceuticals. J.R. Marszalek reported a patent to IACS-010759 issued. K. Takahashi reported personal fees from Celgene during the conduct of the study; and personal fees from Symbio Pharmaceuticals, GSK, and Novartis outside the submitted work. C.D. DiNardo reported personal fees from Agios Pharmaceuticals, Celgene, and AbbVie outside the submitted work. M. Konopleva reported "other" from Amgen, Kisoji, and Reata Pharmaceutical; and grants from AbbVie, Genentech, and Stemline Therapeutics, F. Hoffman La-Roche, Forty Seven, Eli Lilly, Cellectis, Calithera, Ablynx, Agios, Ascentage, Astra Zeneca, Rafael Pharmaceutical, and Sanofi outside the submitted work. In addition, M. Konopleva had a patent to Novartis pending (62/993,166), a patent to Eli Lilly issued, and a patent to Reata Pharmaceutical issued (7,795,305 B2 CDDO). C. Récher reported grants from Celgene, Amgen, Novartis, Jazz, AbbVie, Astellas, MaatPharma, Agios, Daiichi-Sankyo, and Roche; personal fees from Incyte, Macrogenics, Otsuka, Janssen, Pfizer, and Takeda; and non-financial support from Sanofi and Gilead outside the submitted work. No other disclosures were reported.
Figures








References
-
- Alvarez-Calderon, F., Gregory M.A., Pham-Danis C., DeRyckere D., Stevens B.M., Zaberezhnyy V., Hill A.A., Gemta L., Kumar A., Kumar V., et al. 2015. Tyrosine kinase inhibition in leukemia induces an altered metabolic state sensitive to mitochondrial perturbations. Clin. Cancer Res. 21:1360–1372. 10.1158/1078-0432.CCR-14-2146 - DOI - PMC - PubMed
-
- Amatangelo, M.D., Quek L., Shih A., Stein E.M., Roshal M., David M.D., Marteyn B., Farnoud N.R., de Botton S., Bernard O.A., et al. 2017. Enasidenib induces acute myeloid leukemia cell differentiation to promote clinical response. Blood. 130:732–741. 10.1182/blood-2017-04-779447 - DOI - PMC - PubMed
-
- Baccelli, I., Gareau Y., Lehnertz B., Gingras S., Spinella J.-F., Corneau S., Mayotte N., Girard S., Frechette M., Blouin-Chagnon V., et al. 2019. Mubritinib Targets the Electron Transport Chain Complex I and Reveals the Landscape of OXPHOS Dependency in Acute Myeloid Leukemia. Cancer Cell. 36:84–99.e8. 10.1016/j.ccell.2019.06.003 - DOI - PubMed
-
- Bhalla, K., Hwang B.J., Dewi R.E., Ou L., Twaddel W., Fang H.B., Vafai S.B., Vazquez F., Puigserver P., Boros L., and Girnun G.D.. 2011. PGC1α promotes tumor growth by inducing gene expression programs supporting lipogenesis. Cancer Res. 71:6888–6898. 10.1158/0008-5472.CAN-11-1011 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

