STREME: accurate and versatile sequence motif discovery
- PMID: 33760053
- PMCID: PMC8479671
- DOI: 10.1093/bioinformatics/btab203
STREME: accurate and versatile sequence motif discovery
Abstract
Motivation: Sequence motif discovery algorithms can identify novel sequence patterns that perform biological functions in DNA, RNA and protein sequences-for example, the binding site motifs of DNA- and RNA-binding proteins.
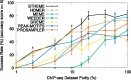
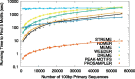
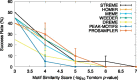
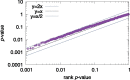
Results: The STREME algorithm presented here advances the state-of-the-art in ab initio motif discovery in terms of both accuracy and versatility. Using in vivo DNA (ChIP-seq) and RNA (CLIP-seq) data, and validating motifs with reference motifs derived from in vitro data, we show that STREME is more accurate, sensitive and thorough than several widely used algorithms (DREME, HOMER, MEME, Peak-motifs) and two other representative algorithms (ProSampler and Weeder). STREME's capabilities include the ability to find motifs in datasets with hundreds of thousands of sequences, to find both short and long motifs (from 3 to 30 positions), to perform differential motif discovery in pairs of sequence datasets, and to find motifs in sequences over virtually any alphabet (DNA, RNA, protein and user-defined alphabets). Unlike most motif discovery algorithms, STREME reports a useful estimate of the statistical significance of each motif it discovers. STREME is easy to use individually via its web server or via the command line, and is completely integrated with the widely used MEME Suite of sequence analysis tools. The name STREME stands for 'Simple, Thorough, Rapid, Enriched Motif Elicitation'.
Availability and implementation: The STREME web server and source code are provided freely for non-commercial use at http://meme-suite.org.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author(s) 2021. Published by Oxford University Press. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures




References
-
- Bailey T.L., Elkan C. (1995) The value of prior knowledge in discovering motifs with MEME. In Proceedings of the Third International Conference on Intelligent Systems for Molecular Biology, July 16–19, 1995, Cambridge, UK, Vol. 3, pp. 21–29. - PubMed
-
- Fisher R.A. (1922) On the interpretation of from contingency tables, and the calculation of p. J. R. Stat. Soc., 85, 87–94.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

